 |
J Circ Biomark 2024; 13: 36-44 ISSN 1849-4544 | DOI: 10.33393/jcb.2024.3146 ORIGINAL RESEARCH ARTICLE |
 |
Altered amino and fatty acids metabolism in Sudanese prostate cancer patients: insights from metabolic analysis
ABSTRACT
Introduction: Prostate cancer (PCa) management presents a multifaceted clinical challenge, intricately linking oncological considerations with cardiovascular health. Despite the recognized importance of lipid metabolism and hypertension in this interwoven relationship, their involvement in PCa development remains partially understood. This study aimed to explore variations in plasma metabolome among Sudanese PCa patients and their associated comorbidities.
Methods: Plasma samples were collected from 50 patients across four hospitals in Sudan and profiled by nuclear magnetic resonance (NMR) spectroscopy. One-dimensional proton NMR spectra were acquired for each sample using standard nuclear Overhauser effect spectroscopy pulse sequence presat on a 500 MHz Bruker Avance III HD NMR spectrometer. Metabolite concentrations were quantified using R scripts developed in-house. Univariate and multivariate analyses were generated in the R software.
Results: Patients were categorized into four distinct metabotypes based on their metabolic profiles, and statistical analyses were conducted to evaluate the significance of observed differences. Our findings revealed high levels of fatty acids, phospholipids, cholesterol, valine, leucine, and isoleucine associated with non-hypertensive patients. In contrast, hypertensive patients were associated with high GlycA and GlycB levels and altered amino acid metabolism.
Conclusion: These findings underscore the intricate interplay between metabolic dysregulation and hypertension in PCa patients. Further research is warranted to elucidate the precise molecular pathways underlying lipid metabolism in PCa and to explore the therapeutic potential of targeting these pathways. In conclusion, our study contributes to a deeper understanding of the metabolic landscape of PCa in Sudanese patients, emphasizing the importance of personalized approaches in cancer management.
Keywords: Africa, Metabolomics, NMR, Prostate cancer, Sudan
Received: May 22, 2024
Accepted: November 20, 2024
Published online: December 16, 2024
This article includes supplementary material
Journal of Circulating Biomarkers - ISSN 1849-4544 - www.aboutscience.eu/jcb
© 2024 The Authors. This article is published by AboutScience and licensed under Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0).
Commercial use is not permitted and is subject to Publisher’s permissions. Full information is available at www.aboutscience.eu
Introduction
Prostate cancer (PCa) poses a significant global health challenge, characterized by its increasing incidence and substantial impact on patient morbidity and mortality (1). With advancements in anticancer therapies leading to improved survival rates, the emergence of cardiovascular toxicities and hypertension (HTN) presents clinicians with intricate management dilemmas (2). Understanding the complex biochemical alterations associated with PCa is vital for advancing diagnostic and therapeutic strategies (3). Metabolomics offers a comprehensive snapshot of the metabolome, the complete set of small molecule metabolites within a biological system, providing insights into dynamic changes occurring in cellular metabolism (4,5). It has emerged as a pivotal tool in elucidating the intricate metabolic changes underlying PCa development and progression (6–9). Particularly, the investigation of altered metabolic pathways involving lipids, fatty acids, and free amino acids holds immense importance in understanding the molecular mechanisms driving PCa pathogenesis (10,11).
Among the plethora of molecular factors implicated in PCa pathogenesis, lipids have emerged as pivotal players, governing diverse cellular processes crucial for tumor progression (12). Emerging evidence suggests a complex interplay between lipid metabolism, HTN, and androgen deprivation therapies (ADTs) in PCa patients, adding further layers of complexity to patient care (13,14). Dysregulated lipid metabolism not only fuels the energy demands of proliferating cancer cells but also contributes to the structural integrity of cellular membranes and facilitates signaling pathways crucial for PCa progression (15). Fatty acids, the building blocks of complex lipids, are intricately involved in various cellular processes, including energy production, membrane synthesis, and signaling modulation (16). Perturbations in fatty acid metabolism have been implicated in PCa pathophysiology, influencing tumor aggressiveness, therapeutic resistance, and disease prognosis (17). Metabolomic studies have unveiled alterations in fatty acid composition and metabolism associated with PCa, highlighting their potential as biomarkers for disease diagnosis and therapeutic targets for intervention (10,18). Moreover, free amino acids play pivotal roles in cellular metabolism, serving as precursors for protein synthesis, energy production, and signaling molecules (19). Alteration in amino acid metabolism has been implicated in PCa progression, influencing cell proliferation, invasion, and metastasis (20,21). Metabolomic profiling has uncovered variations in amino acid levels and metabolism in PCa, offering insights into the relation between metabolic rewiring and oncogenic signaling pathways (8,11).
Most metabolomic PCa studies are conducted predominantly in European and Asian populations. The generalization of findings to other ethnic groups may be limited due to inherent genetic, environmental, and lifestyle differences (22). Therefore, investigating altered metabolic pathways in diverse populations, including those from African regions like Sudan, is crucial for elucidating population-specific variations in PCa biology. Sudan presents a unique demographic landscape characterized by its distinct genetic ancestry, dietary habits, and environmental exposures (23). The region is characterized by a rich diversity highlighted by the extensive presence of major continental African language families, including Niger-Congo (NC), Nilo-Saharan (NS), and Afro-Asiatic (AA) (24). Our study aims to reveal underlying metabolic phenotypes across Sudanese PCa patients to better understand the triggering mechanisms and corresponding tumor heterogeneity.
Material and methods
Study patients and sample collection
The study encompasses a cohort of Sudanese patients diagnosed with PCa. They were primarily treated at the Urology Department of four facilities: Omdrman Teaching Hospital (OTH), Kuwaiti Specialized Hospital (KSH), Soba University Hospital (SUH), and Military Crop (MC). Ethical approval was provided by the National Health Research Ethics Committee of the Sudanese Federal Ministry of Health. The prevalence of comorbidities such as HTN and type 2 diabetes mellitus (T2DM) was documented. Further insights into PCa characteristics including prostate-specific antigen (PSA) levels, biopsy types, perineural invasion (PNI), and disease grading were recorded from Grade 1 to Grade 5. Written consent was obtained from all the participants for tissue and blood examinations. All clinical data were collected regarding their age and clinical history. Five milliliters of blood was collected in Vacuette® ethylenediaminetetraacetic acid (EDTA) tube by medical staff. Blood plasma was separated by centrifugation (1000 g for 10 min at 4°C) and stored at –80°C. Serum PSA level was estimated.
Nuclear magnetic resonance sample preparation and analysis
Plasma samples were thawed at room temperature. An aliquot of 350 μL of a phosphate sodium buffer (70 mM Na2HPO4; 20% (v/v) 2H2O; 6.1 mM NaN3; 4.6 mM sodium 3-trimethylsilyl [2,2,3,3-2H4]-propionate; pH 7.4) was added to 350 μL of each sample. The mixture was homogenized by vortexing for 30 s, before 600 μL of this mixture was transferred into a 5-mm nuclear magnetic resonance (NMR) tube for analysis. One-dimensional (1D) proton (1H)-NMR spectra were acquired on a 500 MHz Bruker Avance III HD NMR spectrometer equipped with a triple-resonance inverse 1H probe head and x, y, z gradient coils. A standard nuclear Overhauser effect spectroscopy (NOESY) pulse sequence presat (noesygppr1d) was used on plasma samples (25). Pooled samples were used as a quality control sample and were included in each batch for qualitative assessment of repeatability by overlaying the raw spectra. The peaks of the identified metabolites were fitted by combining a local baseline and Voigt functions based on the multiplicity of the NMR signal (26). To validate the efficacy of the different deconvolution models, the root-mean-square deviation was determined. The absolute concentration of each metabolite was calculated according to the previously reported equation (27). The number of protons contributing to the unknown signals was imputed to 1. The concentration of carbohydrates was also estimated by considering the equilibrium between their cyclic forms. GlycA and GlycB signals were quantified by integrating the areas between 2.00 and 2.05 ppm and between 2.09 and 2.05 ppm, respectively, above a local baseline, aiming to remove the signal of lipoproteins.
Statistical and data analysis
Statistical analysis and graphical illustrations of the data were generated in the R (version 4.3.2) and R studio (version 1.1.456) software using scripts developed in-house. Wilcoxon rank sum test and Kruskal-Wallis rank sum test were used to compare differences in numerical covariates (e.g., age and metabolite concentration). Fisher’s exact test was used to assess differences between categorical variables (e.g., ethnicity). Spearman’s test was used to calculate the correlation coefficient (rho) between variables. Unsupervised analysis was performed on the metabolic profiles using the KODAMA algorithm (28,29). Hierarchical clustering (Ward linkage) (30) was used on the KODAMA’s output to identify clusters with similar metabolic patterns. Silhouette median value was used to evaluate the optimal number of clusters with the number of possible clusters varying from 2 to 10 (31). The p values less than 0.05 were considered to be significant. To account for multiple tests, a false discovery rate (FDR) of <10% was applied. Linear scaling was used for normalization. Logistic regression models were used to calculate odds ratio (OR) estimates with 95% confidence intervals (CIs) of association between HTN and measured NMR metabolites using ggforestplot R package (32).
Results
Patients’ demographic and clinicopathological characteristics
The participants recruited for this study are representative of the three predominant ethnic groups prevailing in Sudan. The ethnic distribution among the 50 patients included in the study reveals a predominant representation of AA, constituting 48% (n = 24) of the cohort, followed by 34% (n = 17) identified as NC, and 18% (n = 9) as NS. Regarding hospital distribution, KSH accounted for 58% (n = 29) of the total recruited patients, followed by MC at 16% (n = 8), OTH at 14% (n = 7), and SUH at 12% (n = 6). The patients from these three ethnic groups exhibited comparable age distributions and prevalence of disease comorbidities such as HTN and T2DM. No significant differences were observed in PSA levels across ethnic groups. Most of the recruited patients were diagnosed utilizing tissues from transurethral resection of the prostate (TURP), accounting for 68% (n = 34), followed by needle biopsy at 26% (n = 13), prostatectomy at 4% (n = 2), and transurethral laser vaporization of the prostate (TVP) at 2% (n = 1). PNI was observed in 11 tissue samples. Regarding disease grading, 34% of patients were classified as grade 5 PCa (n = 17), 34% as grade 4 (n = 17), 10% as grade 3 (n = 5), 12% as grade 2 (n = 6), and 10% as grade 1 (n = 5). There were no significant differences in clinicopathological parameters observed among patients belonging to the three ethnic groups. A detailed summary of the clinical and demographic characteristics of PCa within the analyzed cohort is presented in Table I.
| Feature | AA (n = 24) | NC (n = 17) | NS (n = 9) | p-Value |
|---|---|---|---|---|
| Hospital | 0.083 | |||
| KSH, n (%) | 16 (66.7) | 10 (58.8) | 3 (33.3) | |
| MC, n (%) | 2 (8.3) | 5 (29.4) | 1 (11.1) | |
| OTH, n (%) | 2 (8.3) | 2 (11.8) | 3 (33.3) | |
| SUH, n (%) | 4 (16.7) | 0 (0.0) | 2 (22.2) | |
| Age (years), median [IQR] | 74.5 [65-77] | 75 [62-78] | 65 [63-71] | 0.124 |
| Hypertension | 0.922 | |||
| No, n (%) | 9 (37.5) | 5 (29.4) | 3 (33.3) | |
| Yes, n (%) | 15 (62.5) | 12 (70.6) | 6 (66.7) | |
| T2DM | 0.321 | |||
| No, n (%) | 12 (50.0) | 12 (70.6) | 4 (44.4) | |
| Yes, n (%) | 12 (50.0) | 5 (29.4) | 5 (55.6) | |
| PSA (ng/mL), median [IQR] | 42.35 [24.4-63] | 54 [37.4-62.1] | 50.5 [44.9-94.2] | 0.238 |
| Biopsy type | 0.858 | |||
| Needle biopsy, n (%) | 8 (33.3) | 3 (17.6) | 2 (22.2) | |
| Prostatectomy, n (%) | 1 (4.2) | 1 (5.9) | 0 (0.0) | |
| TVP, n (%) | 1 (4.2) | 0 (0.0) | 0 (0.0) | |
| TURP, n (%) | 14 (58.3) | 13 (76.5) | 7 (77.8) | |
| PNI | 0.345 | |||
| No, n (%) | 20 (83.3) | 11 (64.7) | 8 (88.9) | |
| Yes, n (%) | 4 (16.7) | 6 (35.3) | 1 (11.1) | |
| Grade | 0.572 | |||
| 1, n (%) | 3 (12.5) | 1 (5.9) | 1 (11.1) | |
| 2, n (%) | 5 (20.8) | 0 (0.0) | 1 (11.1) | |
| 3, n (%) | 1 (4.2) | 3 (17.6) | 1 (11.1) | |
| 4, n (%) | 7 (29.2) | 7 (41.2) | 3 (33.3) | |
| 5, n (%) | 8 (33.3) | 6 (35.3) | 3 (33.3) |
AA = Afro-Asiatic; IQR = interquartile range; KSH = Kuwaiti Specialized Hospital; MC = Military Crop; NC = Niger-Congo; NS = Nilo-Saharan; OTH = Omdurman Teaching Hospital; PNI = perineural invasion; PSA = prostate-specific antigen; SUH = Soba University Hospital; T2DM = type 2 diabetes mellitus; TURP = transurethral resection of prostate; TVP = transurethral laser vaporization of prostate.
Metabolic profiling
NMR spectroscopy was used to profile the plasma metabolome. The metabolic profiles from the three ethnic backgrounds were analyzed (Tab. S1), revealing no significant differences in the metabolic landscape across different ethnicities. However, we reported strong correlations in certain metabolic groups, as shown in Figure 1A. Lipid metabolites encompassing cholesterol, glycerol phospholipid, lipid = CH-CH2-CH = , lipid alpha-CH2, lipid beta-CH2, lipid CH2, lipid CH3, and unsaturated lipid -CH = CH- exhibited distinctive patterns. They showed a significant correlation with the amino acids valine, leucine, and isoleucine. The glutamine was negatively related to glutamate levels. Pyruvate exhibited a negative correlation with glutamate and histidine while displaying a positive correlation with citrate, glycine, and lactate. As expected, the inflammatory NMR markers GlycA and GlycB were negatively correlated with the concentration level of histidine, as reported previously (33). Additionally, GlycA and GlycB were also negatively correlated with the lipid levels.
Metabolic phenotype analysis
The plasma metabolic profile of PCa patients is heterogeneous and shaped by different factors, including inflammation and lipid metabolism. To elucidate metabolic patterns in PCa patients, we employed the unsupervised KODAMA method to analyze quantified NMR metabolite concentrations. By grouping patients with similar metabolic profiles together, four distinct metabolic phenotypes (metabotypes) were identified, as visualized in the heatmap of Figure 2B. The demographic and clinical variations among four identified patient metabotypes are illustrated in Table II. While some differences exist, many parameters do not reach statistical significance among the metabotypes. Significantly divergent HTN prevalence (p-value = 0.028) was observed among the predicted patient groups. Metabotypes A, B, and D exhibited a higher proportion of hypertensive patients (76.2%, 70%, and 80%, respectively) compared to metabotype B, which predominantly comprised non-hypertensive patients (77.8%). The variations in metabolic signature across four metabotypes with corresponding cancer grade and HTN status are represented in Figure 2C.
Through a detailed metabolic comparison illustrated in Table S2, it becomes evident that these metabotypes exhibit distinct metabolic patterns, each characterized by unique compositions of metabolites. Among the 38 NMR metabolites measured, 23 metabolites exhibited significant differences between the predicted metabotypes, each forming a distinct metabolic pattern that characterized the identified metabotypes uniquely (Fig. 2A). The metabotype A is characterized by elevated levels of formate, glutamine, and acetate. In contrast, metabotype B displayed an elevated set of fatty acids and lipid metabolites associated with high levels of some amino acids. Conversely, the metabotype C exhibited higher levels of ascorbate, glutamate, and histidine, while metabotype D showed a distinct pattern of elevated levels of inflammatory biomarkers GlycA and GlycB. These findings indicate unique metabolic signatures associated with each group. Since high circulating cholesterol increases the risk of PCa aggressiveness (15), its level across the four metabotype groups is visualized relative to the HTN status of corresponding patients (Fig. 2B). Additionally, GlycB, serving as an inflammatory biomarker and previously reported to be correlated with a high lipidomic profile (33), also displayed regarding the HTN status of patients from different metabotypes (Fig. 2C).
Association of hypertension with patient characteristics
Our findings indicate that despite a higher proportion of non-hypertensive patients falling under metabotype B, their lipid metabolite levels remain elevated compared to other groups. To uncover potential metabolic patterns associated with hypertension, a thorough examination of demographic, clinical (Tab. S3), and metabolic (Tab. S4) data comparing patients with and without HTN has been performed. Our analysis unveiled significantly elevated levels of PSA in hypertensive patients compared to the non-hypertensive group (p-value = 0.037), remarking the association between hypertension and prostate health. A discrepancy in the T2DM prevalence was observed between hypertensive and non-hypertensive groups (p-value = 0.021). Most hypertensive patients (81.8%) were found to be non-diabetic while 94.1% of non-hypertensive patients presented with T2DM. Moreover, hypertensive patients exhibited significantly higher concentrations of various lipid metabolites compared to their non-hypertensive counterparts. These included unsaturated lipid -CH = CH- (p-value = 0.021), lipid alpha-CH2 (p-value = 0.007), lipid CH2 (p-value = 0.002), lipid beta-CH2 (p-value = 0.02), and glycerol phospholipid (p-value = 0.005), as evidenced by our statistical analysis (Fig. 3, Tab. S4). These findings underscore the intricate metabolic alterations associated with hypertension and the need for further explorations in understanding the pathophysiology of hypertension and its complications in PCa patients.
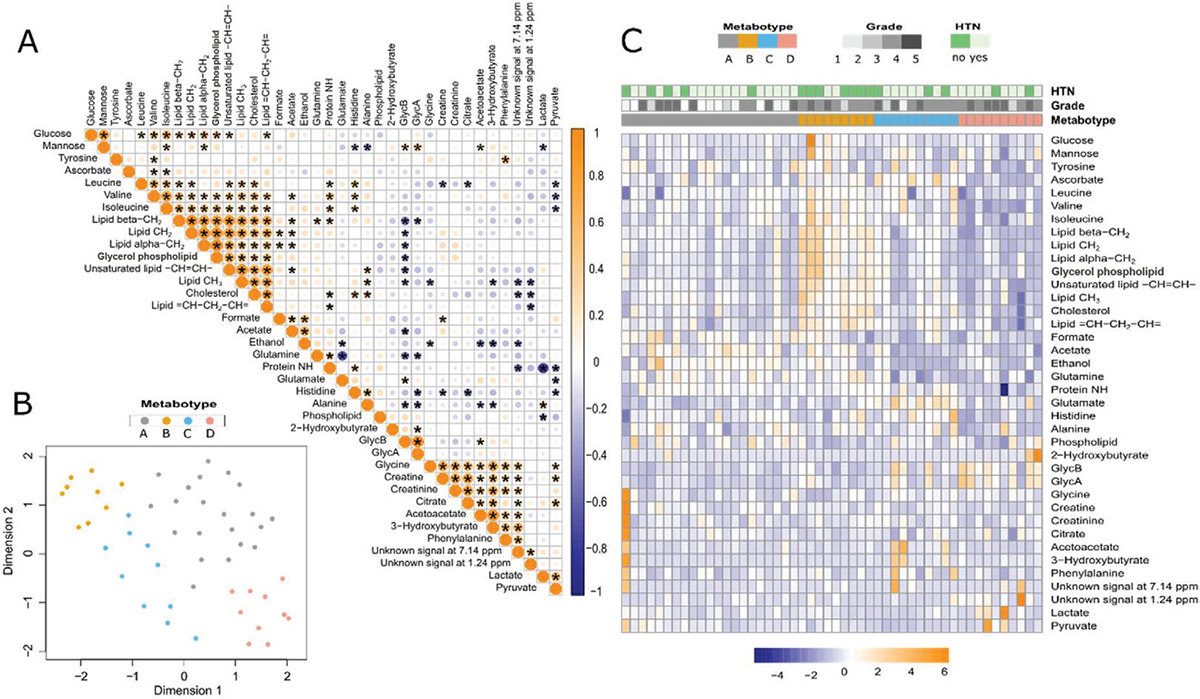
FIGURE 1 - Descriptive analysis for NMR metabolic profile of PCa patients. A) Correlation matrix of Spearman’s rank correlation coefficients between the estimated metabolites. B) KODAMA plot visualizing four metabotypes. C) Heatmap showing the comparison of immune cell profiles across four metabotypes. HTN = hypertension; NMR = nuclear magnetic resonance; PCa = prostate cancer.
| Feature | Metabotype A
(n = 21) |
Metabotype B
(n = 9) |
Metabotype C
(n = 10) |
Metabotype D
(n = 10) |
p-Value |
|---|---|---|---|---|---|
| Hospital | 0.073 | ||||
| KSH, n (%) | 8 (38.1) | 7 (77.8) | 5 (50.0) | 9 (90.0) | |
| MC, n (%) | 4 (19.0) | 0 (0.0) | 3 (30.0) | 1 (10.0) | |
| OTH, n (%) | 5 (23.8) | 2 (22.2) | 0 (0.0) | 0 (0.0) | |
| SUH, n (%) | 4 (19.0) | 0 (0.0) | 2 (20.0) | 0 (0.0) | |
| Ethnicity | 0.342 | ||||
| AA, n (%) | 13 (61.9) | 4 (44.4) | 2 (20.0) | 5 (50.0) | |
| NC, n (%) | 4 (19.0) | 4 (44.4) | 5 (50.0) | 4 (40.0) | |
| NS, n (%) | 4 (19.0) | 1 (11.1) | 3 (30.0) | 1 (10.0) | |
| Age (year), median [IQR] | 75 [65–80] | 70 [63–72] | 71.5 [68-75] | 61 [57.5-74] | 0.237 |
| HTN | 0.027 | ||||
| No, n (%) | 5 (23.8) | 7 (77.8) | 3 (30.0) | 2 (20.0) | |
| Yes, n (%) | 16 (76.2) | 2 (22.2) | 7 (70.0) | 8 (80.0) | |
| T2DM | 0.408 | ||||
| No, n (%) | 14 (66.7) | 3 (33.3) | 6 (60.0) | 5 (50.0) | |
| Yes, n (%) | 7 (33.3) | 6 (66.7) | 4 (40.0) | 5 (50.0) | |
| PSA (ng/mL), median [IQR] | 42.7[31-61.7] | 35.8[24.9-43.7] | 49.0[44-57.5] | 72.8[46.6-91.6] | 0.231 |
| Biopsy type | 0.611 | ||||
| Needle biopsy, n (%) | 4 (19.0) | 2 (22.2) | 4 (40.0) | 3 (30.0) | |
| …Prostatectomy, n (%) | 1 (4.8) | 0 (0.0) | 1 (10.0) | 0 (0.0) | |
| TVP, n (%) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (10.0) | |
| TURP, n (%) | 16 (76.2) | 7 (77.8) | 5 (50.0) | 6 (60.0) | |
| PNI | 0.521 | ||||
| No, n (%) | 17 (81.0) | 8 (88.9) | 8 (80.0) | 6 (60.0) | |
| Yes, n (%) | 4 (19.0) | 1 (11.1) | 2 (20.0) | 4 (40.0) | |
| Grade | 0.056 | ||||
| 1, n (%) | 5 (23.8) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| 2, n (%) | 5 (23.8) | 0 (0.0) | 1 (10.0) | 0 (0.0) | |
| 3, n (%) | 2 (9.5) | 1 (11.1) | 1 (10.0) | 1 (10.0) | |
| 4, n (%) | 2 (9.5) | 6 (66.7) | 5 (50.0) | 4 (40.0) | |
| 5, n (%) | 7 (33.3) | 2 (22.2) | 3 (30.0) | 5 (50.0) |
AA = Afro-Asiatic; HTN = hypertension; IQR = interquartile range; KSH = Kuwaiti Specialized Hospital; MC = Military Crop; NC = Niger-Congo; NS = Nilo-Saharan; OTH = Omdurman Teaching Hospital; PNI = perineural invasion; PSA = prostate-specific antigen; SUH = Soba University Hospital; T2DM = type2 diabetes mellitus; TURP = transurethral resection of prostate; TVP = transurethral laser vaporization of prostate.
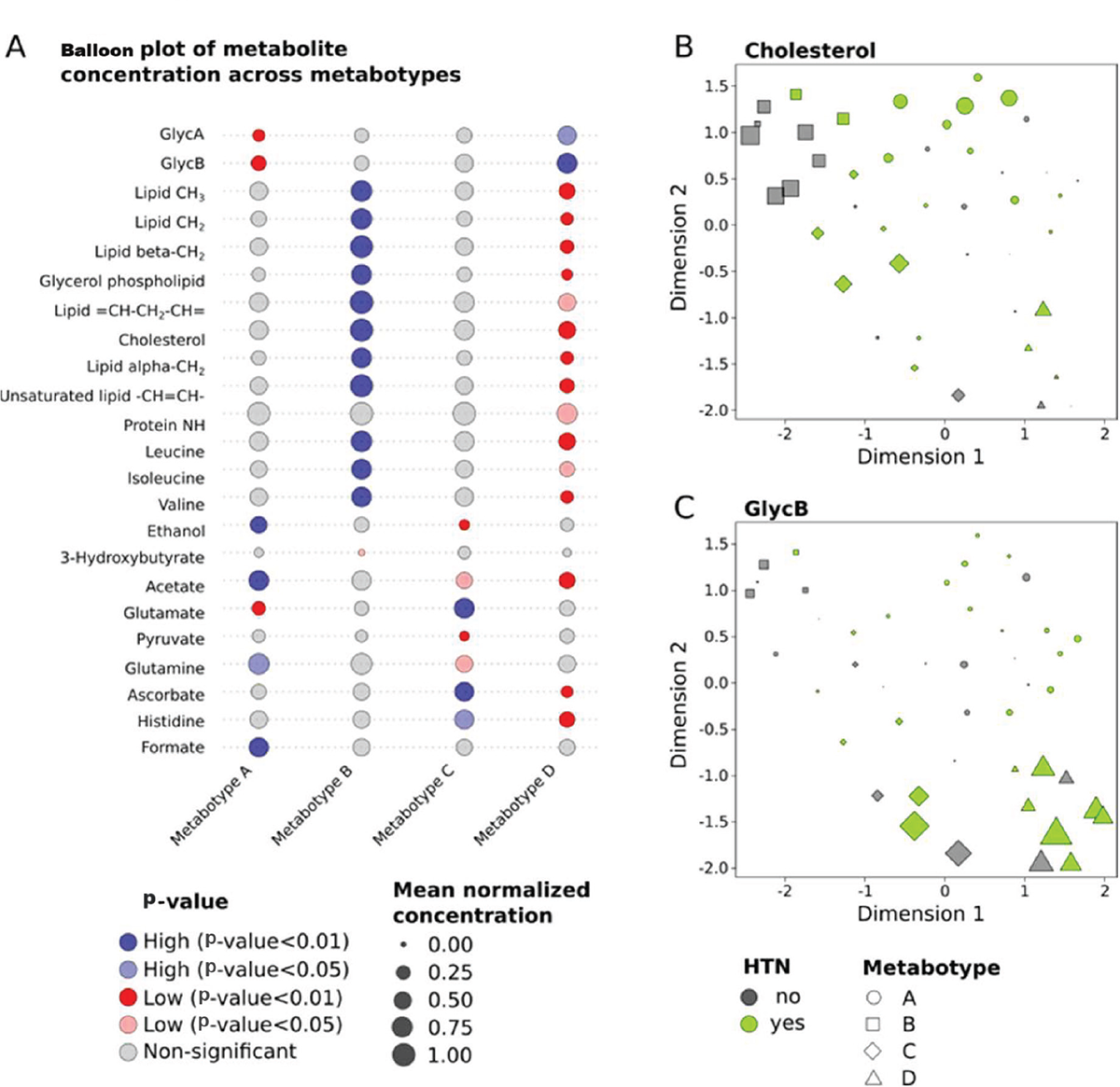
FIGURE 2 - Variations in significant metabolites across four predicted metabotypes. A) Balloon plot representing the different metabolite concentrations across the four metabotypes. The KODAMA plots illustrate the distribution of cholesterol (B) and GlycB concentrations (C), with concentrations represented by varying point sizes across four distinct metabotypes. HTN = hypertension.
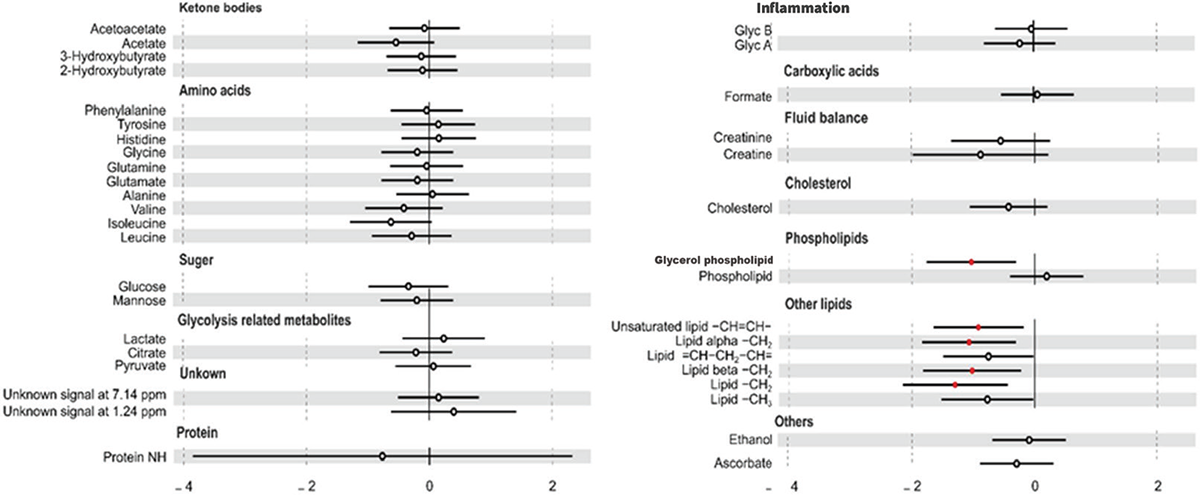
FIGURE 3 - Association between hypertension (HTN) and metabolite concentrations. Odds ratio for HTN per 1-standard deviation increment in metabolite concentration with 95% confidence interval (CI). The red circle represents significant values.
Discussion
Despite the lack of specific metabolomic studies of PCa in Sudanese patients, this research tries to investigate the metabolic profile within the Sudanese context. Mostly, Sudanese PCa patients frequently present with late-stage diagnoses characterized by elevated PSA levels and Gleason scores (34). The diverse ethnic composition of our study, with a predominant representation of ethnicities such as AA, NC, and NS, mirrors the demographic diversity typical of this African region (25). Metabolic stratification of PCa patients using machine learning techniques offers a promising approach to elucidate the disease’s heterogeneity and enhance personalized treatment strategies. By analyzing metabolomic profiles from plasma samples, machine learning algorithms can categorize patients into distinct metabolic subtypes based on their unique metabolic signatures. Several studies have showcased the feasibility and potential of this approach. For instance, Cacciatore et al (2021) utilized KODAMA, a clustering algorithm to stratify PCa patients into different metabotypes using metabolomic profiling data (33). Their findings highlighted distinct inflammatory metabolic patterns associated with PCa aggressiveness in South African patients.
In our study, we applied KODAMA to categorize Sudanese PCa patients based on their NMR-measured metabolic data, identifying four distinct metabolic groups. These groups differed in PCa grade distribution and exhibited significant variability in hypertension prevalence. An increased level of lipid metabolites, including fatty acids, cholesterol, and glycerol phospholipids, was observed in patients within the second metabotype (metabotype B). This metabolic signature revealed a potential link between lipid metabolism and the clinical characteristics of PCa patients grouped within this group. Unlike other metabotypes, this group is dominated by non-hypertensive patients, which implies another factor that can interfere with the recorded raised lipid metabolites. Previous research implicated dysregulated lipid metabolism in cancer progression, including PCa (35–37). Accumulation of phospholipids and cholesterol in the circulation interacts with androgens and the immune system and triggers PCa development (38). The extended utilization of ADT is recognized for inducing alterations in the lipid profile both during and following treatment (39,40). It may be potentially contributing to the enhanced lipid biosynthesis in metabotype B group. Furthermore, our analysis also revealed a positive correlation between estimated lipid metabolites and certain amino acids, particularly valine, leucine, and isoleucine, which are classified as branched-chain amino acids (BCAAs). These BCAAs play pivotal roles in crucial cellular processes such as protein synthesis, energy production, and cellular signaling (41). The dysregulation of BCAA metabolism has been implicated in the pathogenesis of various cancers, including PCa (42). The genes encoding these amino acids have been reported to be expressed at higher levels in PCa tumor tissue compared to non-transformed tissue (43). Previous studies have illuminated the relation between lipid and amino acid metabolism in various cancer types (44,45). Specifically, cancer cells, including those of PCa, often exhibit heightened demands for lipid synthesis and fatty acid oxidation to fuel their growth and meet their energy requirements, while simultaneously orchestrating the transport and metabolic pathways of amino acids to sustain proliferation and ensure survival (46). Aside from BCAAs, our results also highlighted variations in the concentrations of other metabolites such as glutamine, glutamate, histidine, and pyruvate across the detected metabotype groups. The abnormal metabolism of amino acids in PCa cells often manifests as a disruption in glutamic acid metabolism, which is synthesized from glutamine to fulfill the demands of growth and proliferation (47). Plasma samples from PCa patients have been shown to exhibit elevated levels of valine, glutamine, creatine, tyrosine, phenylalanine, histidine, and 3-methylhistidine (48). Increased levels of BCAAs, glutamate, and a reduction in the levels of glycine, dimethylglycine, fumarate, and 4-imidazole-acetate have been identified as discriminative factors between PCa patients and those with benign prostatic hyperplasia (49). Lipid oxidation might be initiated by glycosylation of phospholipids leading to formation of advanced glycation end products. This pathway might increase the severity of inflammatory diseases, and the initiation and progression of malignant tumors (50). GlycA and GlycB are identifiable as two distinct signals in NMR spectroscopy that indicate the glycation status of the most abundant acute-phase inflammatory proteins, reflecting the patient inflammatory status (51). From our results, there was increased concentrations of GlycA and GlycB in the fourth metabotype (metabotype D) associated with low levels of lipid metabolites. Consistent with our results, a previous study reported a similar PCa metabotype, characterized by a high level of GlycB and reduced concentration of lipids (33). The possible origin of this metabotype is still under investigation.
Conclusion
This study sheds light on the metabolic landscape of PCa in Sudanese patients, emphasizing the potential of metabolomics in enhancing personalized treatment strategies. The identified metabolic subtypes using machine learning techniques provide valuable insights into disease heterogeneity, guiding targeted interventions. The associations between lipid metabolites and PCa characteristics underscore the importance of metabolic dysregulation in disease progression. Further research into the interplay between lipid and amino acid metabolism, informed by our findings and previous studies, promises to deepen our understanding of PCa pathogenesis and identify novel therapeutic targets.
Limitations and future directions
While the study provided valuable insights into the metabolic variations among PCa patients with comorbidities like hypertension, this study has some limitations that should be considered. The relatively small sample size (50 patients) limits the generalizability of the findings, and the focus on a specific population in Sudan may not fully capture metabolic variations across different regions or ethnic groups. Finally, while NMR spectroscopy was useful for metabolomic profiling, it may have missed other relevant metabolites that more sensitive techniques like mass spectrometry could detect. Despite these limitations, the study offers practical insights identifying distinct metabolic phenotypes in PCa patients, particularly concerning hypertension. The findings may also contribute to the development of biomarkers for early detection and risk stratification, particularly in patients with metabolic comorbidities like hypertension. Further research could explore these metabolic pathways to inform targeted therapies.
Acknowledgments
We would like to express our deep and sincere gratitude and appreciation to all urology and histopathology surgeons, clinicians, and technical staff for their efforts in performing this study.
Disclosures
Conflicts of Interest: The authors declare no conflicts of interest.
Financial support: The research was funded by the International Centre for Genetic Engineering and Biotechnology (L.F.Z. and S.C.), and the UN Technology Bank for Least Developed Countries, TWAS and ICGEB under South-North and South-South Fellowships (D.A.).
Informed consent statement: Informed consent was taken from all the participants.
Author contributions: Conceptualization, D.A., S.C., and L.F.Z.; methodology, S.C.; formal analysis, D.A., E.A.A.S., S.C.; resources, D.A., E.A.A.M., H.E.A.B.A., A.M.I., S.C., and L.F.Z.; clinical data curation, D.A., E.A.A.M., H.E.A.B.A., A.M.I.; writing – original draft preparation, D.A. and E.A.A.S.; writing – review and editing, S.C. and L.F.Z.; visualization, E.A.A.S.; supervision, S.C. and L.F.Z.; project administration, S.C. and L.F.Z.; funding acquisition, D.A., S.C., and L.F.Z. All authors have read and agreed to the published version of the manuscript.
Institutional review board statement: Specimen collection and research contents were examined and approved by the National Health Research Ethics Committee of the Sudanese Federal Ministry of Health (6-9-2021). The study was performed in accordance with the Declaration of Helsinki.
Data availability statement: All data produced in the present study are available upon reasonable request to the authors. NMR spectra were deposited at the MetaboLight database.
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394-424. CrossRef PubMed
- 2. Choi Y, Park B, Jeong BC, et al. Usefulness of early extracorporeal shock wave lithotripsy in colic patients with ureteral stones. Korean J Urol. 2015;56(12):853-859. CrossRef PubMed
- 3. Wasim S, Lee SY, Kim J. Complexities of prostate cancer. Int J Mol Sci. 2022;23(22):14257. CrossRef PubMed
- 4. Danzi F, Pacchiana R, Mafficini A, et al. To metabolomics and beyond: a technological portfolio to investigate cancer metabolism. Signal Transduct Target Ther. 2023;8(1):137. CrossRef PubMed
- 5. Li Z, Zhang H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell Mol Life Sci. 2016;73(2):377-392. CrossRef PubMed
- 6. Kdadra M, Höckner S, Leung H, Kremer W, Schiffer E. Metabolomics biomarkers of prostate cancer: a systematic review. Diagnostics (Basel). 2019;9(1):21. CrossRef PubMed
- 7. Vandergrift LA, Decelle EA, Kurth J, et al. Metabolomic prediction of human prostate cancer aggressiveness: magnetic resonance spectroscopy of histologically benign tissue. Sci Rep. 2018;8(1):4997. CrossRef PubMed
- 8. Struck-Lewicka W, Kordalewska M, Bujak R, et al. Urine metabolic fingerprinting using LC-MS and GC-MS reveals metabolite changes in prostate cancer: a pilot study. J Pharm Biomed Anal. 2015;111:351-361. CrossRef PubMed
- 9. Ren S, Shao Y, Zhao X, et al. Integration of metabolomics and transcriptomics reveals major metabolic pathways and potential biomarker involved in prostate cancer. Mol Cell Proteomics. 2016;15(1):154-163. CrossRef PubMed
- 10. Yang M, Ayuningtyas A, Kenfield SA, et al. Blood fatty acid patterns are associated with prostate cancer risk in a prospective nested case-control study. Cancer Causes Control. 2016;27(9):1153-1161. CrossRef PubMed
- 11. Sroka WD, Boughton BA, Reddy P, et al. Determination of amino acids in urine of patients with prostate cancer and benign prostate growth. Eur J Cancer Prev. 2017;26(2):131-134. CrossRef PubMed
- 12. Li J, Ren S, Piao HL, et al. Integration of lipidomics and transcriptomics unravels aberrant lipid metabolism and defines cholesteryl oleate as potential biomarker of prostate cancer. Sci Rep. 2016;6(1):20984. CrossRef PubMed
- 13. Hahn AW, Thoman W, Koutroumpakis E, et al. Cardiometabolic healthcare for men with prostate cancer: an MD Anderson Cancer Center experience. Cardiooncology. 2023;9(1):33. CrossRef PubMed
- 14. Zhang H, Zhou Y, Xing Z, Sah RK, Hu J, Hu H. Androgen metabolism and response in prostate cancer anti-androgen therapy resistance. Int J Mol Sci. 2022;23(21):13521. CrossRef PubMed
- 15. Zeković M, Bumbaširević U, Živković M, Pejčić T. Alteration of lipid metabolism in prostate cancer: multifaceted oncologic implications. Int J Mol Sci. 2023;24(2):1391. CrossRef PubMed
- 16. Koundouros N, Poulogiannis G. Reprogramming of fatty acid metabolism in cancer. Br J Cancer. 2020;122(1):4-22. CrossRef PubMed
- 17. Minas TZ, Lord BD, Zhang AL, et al. Circulating trans fatty acids are associated with prostate cancer in Ghanaian and American men. Nat Commun. 2023;14(1):4322. CrossRef PubMed
- 18. Noriega Landa E, Quaye GE, Su X, et al. Urinary fatty acid biomarkers for prostate cancer detection. PLoS One. 2024;19(2):e0297615. CrossRef PubMed
- 19. Lieu EL, Nguyen T, Rhyne S, Kim J. Amino acids in cancer. Exp Mol Med. 2020;52(1):15-30. CrossRef PubMed
- 20. Strmiska V, Michalek P, Eckschlager T, et al. Prostate cancer-specific hallmarks of amino acids metabolism: towards a paradigm of precision medicine. Biochim Biophys Acta Rev Cancer. 2019;1871(2):248-258. CrossRef PubMed
- 21. Schcolnik-Cabrera A, Juárez-López D. Dual contribution of the mTOR pathway and of the metabolism of amino acids in prostate cancer. Cell Oncol (Dordr). 2022;45(5):831-859. CrossRef PubMed
- 22. Hinata N, Fujisawa M. Racial differences in prostate cancer characteristics and cancer-specific mortality: an overview. World J Mens Health. 2022;40(2):217-227. CrossRef PubMed
- 23. Ibrahim ME. Genetic diversity of the Sudanese: insights on origin and implications for health. Hum Mol Genet. 2021;30(R1):R37-R41. CrossRef PubMed
- 24. Greenberg JH. The languages of Africa. Bloomington, IN: Indiana University Press 1963. Online (Accessed May 2024)
- 25. Nagana Gowda GA, Raftery D. Analysis of plasma, serum, and whole blood metabolites using 1H NMR spectroscopy. Methods Mol Biol 2019;2037:17-34. CrossRef PubMed
- 26. Marshall I, Higinbotham J, Bruce S, Freise A. Use of Voigt lineshape for quantification of in vivo 1H spectra. Magn Reson Med. 1997;37(5):651-657. CrossRef PubMed
- 27. Serkova N, Fuller TF, Klawitter J, Freise CE, Niemann CU. H-NMR-based metabolic signatures of mild and severe ischemia/reperfusion injury in rat kidney transplants. Kidney Int. 2005;67(3):1142-1151. CrossRef PubMed
- 28. Cacciatore S, Tenori L, Luchinat C, Bennett PR, MacIntyre DA. KODAMA: an R package for knowledge discovery and data mining. Bioinformatics. 2017;33(4):621-623. CrossRef PubMed
- 29. Zinga MM, Abdel-Shafy E, Melak T, et al. KODAMA exploratory analysis in metabolic phenotyping. Front Mol Biosci. 2023;9:1070394. CrossRef PubMed
- 30. Reynolds AP, Richards G, de la Iglesia B, Rayward-Smith VJ. Clustering rules: a comparison of partitioning and hierarchical clustering algorithms. J Math Model Algorithms. 2006;5(4):475-504. CrossRef
- 31. Rousseeuw PJ. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math. 1987;20:53-65. CrossRef
- 32. Akbaraly T, Würtz P, Singh-Manoux A, et al. Association of circulating metabolites with healthy diet and risk of cardiovascular disease: analysis of two cohort studies. Sci Rep. 2018;8(1):8620. CrossRef PubMed
- 33. Cacciatore S, Wium M, Licari C, et al. Inflammatory metabolic profile of South African patients with prostate cancer. Cancer Metab. 2021;9(1):29. CrossRef PubMed
- 34. Taha SM, Weng HY, Mohammed MEI, Osman YM, Mohammed SI, Abuidris DO (2020). Prostate cancer clinical characteristics and outcomes in Central Sudan. Ecancermedicalscience. 2020;14:1116. CrossRef
- 35. Scaglia N, Frontini-López YR, Zadra G. Prostate cancer progression: as a matter of fats. Front Oncol. 2021;11:719865. CrossRef PubMed
- 36. Wu X, Daniels G, Lee P, Monaco ME. Lipid metabolism in prostate cancer. Am J Clin Exp Urol. 2014;2(2):111-120. PubMed
- 37. Zhang Z, Wang W, Kong P, et al. New insights into lipid metabolism and prostate cancer (Review). Int J Oncol. 2023;62(6):1-13. CrossRef PubMed
- 38. Siltari A, Syvälä H, Lou YR, Gao Y, Murtola TJ. Role of lipids and lipid metabolism in prostate cancer progression and the tumor’s immune environment. Cancers (Basel). 2022;14(17):4293. CrossRef PubMed
- 39. Wolny-Rokicka E, Tukiendorf A, Wydmański J, Ostrowska M, Zembroń-Łacny A. Lipid status during combined treatment in prostate cancer patients. Am J Mens Health. 2019;13(5):1557988319876488. CrossRef PubMed
- 40. Zadra G, Loda M. When fat goes down, prostate cancer is on the ropes. Mol Cell Oncol. 2019;6(3):1595308. CrossRef PubMed
- 41. Xu H, Wang X, Xu X, et al. Association of plasma branched-chain amino acid with multiple cancers: a mendelian randomization analysis. Clin Nutr. 2023;42(12):2493-2502. CrossRef PubMed
- 42. Dereziński P, Klupczynska A, Sawicki W, Pałka JA, Kokot ZJ. Amino acid profiles of serum and urine in search for prostate cancer biomarkers: a pilot study. Int J Med Sci. 2017;14(1):1-12. CrossRef PubMed
- 43. Samaržija I, Trošelj KG, Konjevoda P. Prognostic significance of amino acid metabolism-related genes in prostate cancer retrieved by machine learning. Cancers (Basel). 2023;15(4):1309. CrossRef PubMed
- 44. Pardo-Rodriguez D, Santamaría-Torres M, Salinas A, et al. Unveiling disrupted lipid metabolism in benign prostate hyperplasia, prostate cancer, and metastatic patients: insights from a Colombian nested case-control study. Cancers (Basel). 2023;15(22):5465. CrossRef PubMed
- 45. Ahmad F, Cherukuri MK, Choyke PL. Metabolic reprogramming in prostate cancer. Br J Cancer. 2021;125(9):1185-1196. CrossRef PubMed
- 46. Chen L, Xu YX, Wang YS, Zhou JL. Lipid metabolism, amino acid metabolism, and prostate cancer: a crucial metabolic journey. Asian J Androl. 2024;26(2):123-134. CrossRef PubMed
- 47. Stepka P, Vsiansky V, Raudenska M, Gumulec J, Adam V, Masarik M. Metabolic and amino acid alterations of the tumor microenvironment. Curr Med Chem. 2021;28(7):1270-1289. CrossRef PubMed
- 48. Lécuyer L, Victor Bala A, Demidem A, et al. NMR metabolomic profiles associated with long-term risk of prostate cancer. Metabolomics. 2021;17(3):32. CrossRef PubMed
- 49. Pérez-Rambla C, Puchades-Carrasco L, García-Flores M, Rubio-Briones J, López-Guerrero JA, Pineda-Lucena A. Non-invasive urinary metabolomic profiling discriminates prostate cancer from benign prostatic hyperplasia. Metabolomics. 2017;13(5):52. CrossRef PubMed
- 50. Schröter D, Höhn A. Role of advanced glycation end products in carcinogenesis and their therapeutic implications. Curr Pharm Des. 2018;24(44):5245-5251. CrossRef PubMed
- 51. Moreno-Vedia J, Rosales R, Ozcariz E, et al. Triglyceride-rich lipoproteins and glycoprotein A and B assessed by 1H-NMR in metabolic-associated fatty liver disease. Front Endocrinol (Lausanne). 2022;12:775677. CrossRef PubMed