 |
J Circ Biomark 2024; 13: 1-6 ISSN 1849-4544 | DOI: 10.33393/jcb.2024.2636 ORIGINAL RESEARCH ARTICLE |
 |
Detection of PSMA expression on circulating tumor cells by blood-based liquid biopsy in prostate cancer
ABSTRACT
Background: For patients with mCRPC, PSMA-targeted radioligand treatment has significantly improved the clinical outcome. A blood-based liquid biopsy assay for recognizing PSMA protein expression on circulating tumor cells may be beneficial for better informing therapeutic decision-making and identifying the patients most likely to benefit from PSMA-targeted radioligand therapy.
Methods: Using high-throughput imaging and digital AI pathology algorithms, a four-color immunofluorescence assay has been developed to find PSMA protein expression on CTCs on a glass slide. Cell line cells (LNCaP/PC3s/22Rv1) spiked into healthy donor blood were used to study the precision, specificity, sensitivity, limit of detection, and overall accuracy of the assay. Clinical validation and low-pass whole-genome sequencing were performed in PSMA-PET-positive patients with high-risk mCRPC (N = 24) utilizing 3 mL of blood.
Results: The PSMA CTC IF assay achieved analytical specificity, sensitivity, and overall accuracy above 99% with high precision. In the clinical validation, 76% (16/21) of the cases were PSMA positive with CTC heterogeneity, and 88% (21/24) of the patients contained at least one conventional CTC per milliliter of blood. Thirty-six low-pass-sequenced CTCs from 11 individuals with mCRPC frequently exhibited copy number increases in AR and MYC and losses in RB1, PTEN, TP53, and BRCA2 locus.
Conclusions: The analytical validation utilizing Epic Sciences’ liquid biopsy CTC platform demonstrated the potential to detect PSMA protein expression in CTCs from patients with mCRPC. This assay is positioned as an effective research tool to evaluate PSMA expression, heterogeneity, and therapeutic response in many ongoing clinical studies to target tumors that express PSMA.
Keywords: Circulating tumor cells, Liquid biopsy, mCRPC, Prostate-specific membrane antigen (PSMA)
Received: July 18, 2023
Accepted: January 8, 2024
Published online: February 21, 2024
Journal of Circulating Biomarkers - ISSN 1849-4544 - www.aboutscience.eu/jcb
© 2024 The Authors. This article is published by AboutScience and licensed under Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0).
Commercial use is not permitted and is subject to Publisher’s permissions. Full information is available at www.aboutscience.eu
Introduction
A type II transmembrane glycoprotein receptor called prostate-specific membrane antigen (PSMA) is increased in metastatic prostate malignancies and may increase tumor dedifferentiation and treatment resistance (1). The biological basis for PSMA expression in prostate cancer is still unknown; however, research suggests that it plays a role in PI3K-Akt signaling through glutamate signaling (2). Conventional imaging techniques, including magnetic resonance imaging (MRI), computerized tomography (CT), and bone scans, have poor sensitivity and specificity for detecting tumor lesions, especially when prostate-specific antigen (PSA) levels are low. Therefore, PSMA can be used as a target for imaging with radioligands like 68Ga-PSMA-11 and 18F-DCFPyL, which the Food and Drug Administration (FDA) recently approved for PSMA-targeted positron emission tomography (PET) imaging in prostate cancer (3). PSMA is also an established therapeutic target for advanced prostate cancer based on the prospective, phase III VISION trial (NCT03511664) of 177Lu-PSMA-617 radioligand therapy (RLT) that demonstrated improved overall survival (OS) and progression-free survival (PFS) compared to standard of care (SoC) in PSMA-positive advanced metastatic castration-resistant prostate cancer (mCRPC) (4). However, a variety of biological factors, including the stage of the illness (sensitive vs. therapy resistant), the type of treatment, the timing, and geography, may have an impact on PSMA expression in prostate cancer (5).
Analysis of circulating tumor cells is one of the liquid biopsy approaches. Cancer cells known as circulating tumor cells (CTCs) escape from tumors and travel to other organs such as the bones, liver, and lungs via the blood circulation or lymphatic system (6). CTCs are trustworthy, constant, independent predictors and prognostic biomarkers of disease response, PFA, and OS because they directly correlate with systemic tumor burden (7,8). Therefore, it would be advantageous to have a meaningful CTC-based diagnostic and predictive biomarker test that can assist clinical decision-making regarding the management of men with prostate cancer. Furthermore, CTC detection holds enormous potential in numerous clinical applications including cancer screening, monitoring treatment response, diagnosis and prognosis evaluation, target identification, and understanding cancer metastasis and emergence of drug resistance (6,9).
Monitoring the level of PSMA protein expression on CTCs as part of a liquid biopsy offers a noninvasive approach that could be used in conjunction with or in lieu of standard PSMA-PET imaging, which itself presents challenges due to variable access and high cost. Clinically, PSMA CTC profiling is being investigated as a potential prognostic and pharmacodynamic biomarker in patients with mCRPC.
Clinical results in men with mCRPC have been markedly improved by PSMA-targeted radioligand treatment (10,11). Detecting PSMA protein expression on CTCs using a liquid biopsy approach may help to improve therapeutic choices and pinpoint the individuals most likely to benefit from PSMA-targeted RLT in prostate cancer (7-13). In this article, we describe the development and validation of a PSMA CTC assay, as well as the application of single-cell CTC sequencing technology to evaluate deoxyribonucleic acid (DNA) copy number changes and CTC heterogeneity in late-stage high-risk men with mCRPC.
Materials and methods
Cell line and culture
To produce lab-derived (LD) slides, multiple distinct cell line cells (CLCs; LNCaP, PC3, 22Rv1, MDA-MB-231, MDA-MB-453, and DU145) with various PSMA expression levels were spiked into healthy blood. Using sterile procedures and RPMI cell line culture media, all CLCs were grown at Epic Sciences. CLCs were then trypsinized, suspended at single-cell levels, numbered, and spiked into healthy donor (HD) blood samples obtained in a Streck BCT container at the proper CLC concentrations in order to give the CLC for assay development and later assay validation (18).
Epic immunofluorescence PSMA assay
The PSMA tyramide signal amplification (TSA) CTC assay was developed utilizing three different prostate cancer cell lines, including LNCaP (high PSMA expression), 22Rv1 (low PSMA expression), and PC3 (no PSMA expression). These CLCs were spiked into healthy blood samples (1:10,000 CLC/white blood cell [WBC] dilutions) to create LD CLC slides for the PSMA assay development and clinical validation. Subsequently, on a proprietary glass slide, three million intact nucleated blood cells were deposited and frozen at −80°C. In order to identify intact nucleated, biomarker-positive, and negative CTC cells, these frozen slides were defrosted and utilized for immunofluorescent staining with a range of antibodies against cytokeratins, CD45, PSMA, and DAPI. Then, CTCs were detected on immunostained glass slides from patient blood samples using a high-throughput Pyxis imaging system with digital pathology artificial intelligence algorithms. In this four-color PSMA CTC assay, PSMA expression was measured on individual CLC, and CTCs recognized as traditional CTCs (CK+, CD45−, and intact DAPI nuclei) with typical CTC morphological characteristics utilizing an anti-PSMA (Abcam, ERP6253) rabbit monoclonal antibody, as illustrated in Figure 1 (14,18). cRatio, which is the ratio of a cell’s intensity on a specific channel divided by the median cell intensities over all frames in that channel, was applied to determine the CK expression and PSMA protein expression cutoffs (>3 and >6, respectively).
Analytical validation
We computed the PSMA protein biomarker expression’s subcellular localization and assessed the PSMA CTC assay’s limit of detection (LOD), specificity, sensitivity, precision, and overall accuracy. For the accuracy and precision experiments, three independent stain runs for different LD slide lots were examined. To evaluate the specificity of the PSMA TSA assay, three technical replicates each of the PSMA expression negative cell lines, including MDA-MB-231, MDA-MB-453, and DU145, plus HD slides were evaluated, together with two process controls (one positive and one negative). The analytical sensitivity of the PSMA TSA assay was assessed using 14 technical replicates of the LNCaP slide, each with a different number of spike-in CLCs. Additionally, a design of experiments technique was utilized with eight stain repetitions to evaluate the accuracy of the PSMA TSA assay. Furthermore, eight technical replicates and two process control slides were stained to evaluate the PSMA TSA assay’s accuracy. Finally, to evaluate the PSMA TSA assay’s analytical LOD, 24 LD slides containing four different CLC/WBC dilution ratios (1, 10, 30, and 300 CLCs/3E6; six slides each) were evaluated by immunofluorescence (IF) utilizing the optimized PSMA TSA assay. The LOD was determined by analyzing the linearity between the expected and observed numbers of CLCs for each group of slides as well as the lowest number of CLCs identified. The CTC candidates were manually checked by two independent, qualified individuals from Epic Sciences. “CTC per mL” of blood was used to represent the concentration of all CTCs and PSMA-expressing CTCs for each patient (Fig. 1).
Clinical validation and low-pass whole-genome sequencing
A cohort of 24 men with mCRPC and avid PSMA expression confirmed by PSMA-PET/CT scans underwent CTC sampling for clinical validation of the PSMA CTC assay using 3 mL blood samples. Additionally, 36 CTCs from 11 of these men were selected to undergo single-cell low-pass whole-genome sequencing for copy number alteration analysis, as shown in Figure 1 (15). The single CTCs were manually isolated using microcapillaries. They were whole-genome amplified using the SeqPlex™ Enhanced DNA Amplification Kit (SeqXE) from Sigma-Aldrich. The amplified products were made into sequencing libraries and sequenced on an Illumina NextSeq 500 to generate paired-end 150-bp reads. The sequencing data was analyzed using a copy number analysis pipeline that Epic developed (16). Briefly, the sequencing reads were aligned to the hg38 human reference; the low-quality alignments were excluded. The remaining reads were binned, scaled, and corrected for known biases (such as GC bias). The bin counts were then used for copy number variation (CNV) calling using a z-score statistic, which quantifies the amount of deviation in the bin counts of a given cell relative to a population of normal cells.
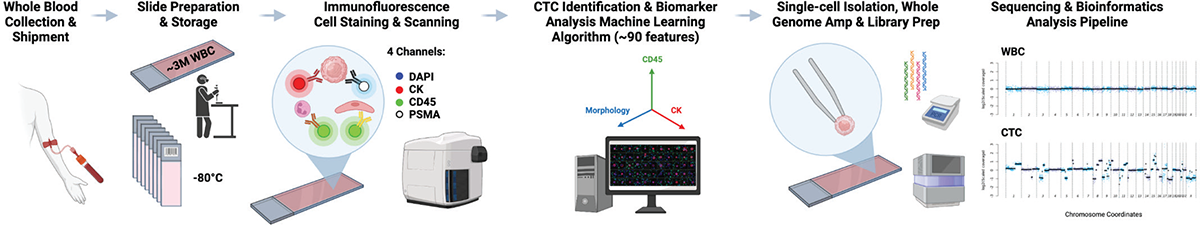
FIGURE 1 - CTC enumeration, biomarker detection, cell picking, and single-cell genomic analysis workflow. CTC = circulating tumor cell.
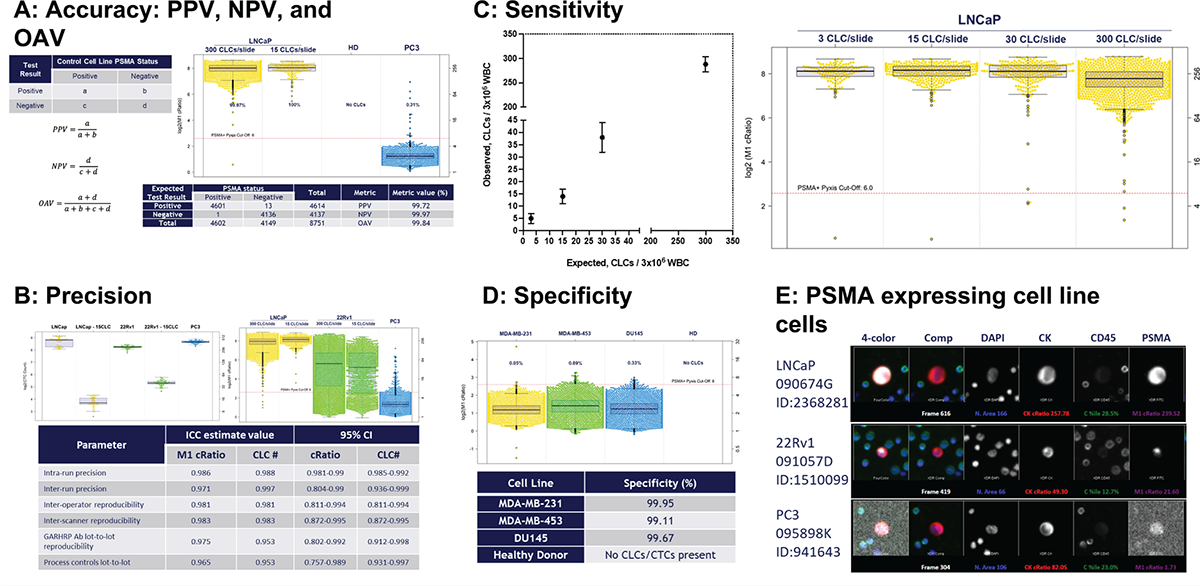
FIGURE 2 - PSMA assay development. A) Denoting the accuracy (PPV, NPV, and OAV) during PSMA assay development. The PSMA assay achieved PPV, NPV, and OAV of over 99%. B) The overall precision of the PSMA assay was ICC 0.98. C) Sensitivity: the lower spike-in concentration in which 1 PSMA+ CLC was detected 90% of the time and the LOD was 1 CLC when 3 CLCs were spiked into the blood sample prior to slide deposition. D) Specificity for the PSMA TSA assay was >99%. E) Gallery of PSMA-expressing CLCs (LNCaP, 22Rv1, and PC3). CLC = cell line cell; ICC = intra-class correlation; LOD = limit of detection; NPV = negative predictive value; OAV = overall accuracy value; PPV = positive predictive value; PSMA = prostate-specific membrane antigen; TSA = tyramide signal amplification.
Results
PSMA assay development utilizing Epic Sciences’ CTC platform
As part of technical feasibility, the development of the PSMA assay was conducted using a multistep approach that included antibody discovery, cell line screening, and the creation of LD slides. Based on a total of 4,614 PSMA-positive and 4,137 PSMA-negative cells, the assay had over 99% positive predictive value, negative predictive value, and overall accuracy value (Fig. 2A). Furthermore, precision was assessed across a variety of metrics, including lot-to-lot reproducibility and process control lot-to-lot repeatability, and it demonstrated high precision (>0.97). To further assess the precision across all intra-run, inter-run, operator, scanner, lot-to-lot repeatability, and process control lots, intra-class correlation (ICC) was calculated; the average result was 0.976 (Fig. 2B). The lower LOD was defined to be the lowest spike-in concentration in which at least one PSMA+ CLC was detected 90% of the time, and the LOD at where 1 CLC/million was spiked into the blood sample prior to slide deposition (Fig. 2C). The specificity for the PSMA TSA assay was calculated based on PSMA-negative, including MDA-MB-231, MDA-MB-453, and DU145, CLCs, which was over 99% (Fig. 2D). A gallery of PSMA-expressing CLCs from LNCaP, 22Rv1, and PC3 are shown in Figure 2E.
PSMA CTC protein expression detection in mCRPC patients
In a cohort of 24 patients with PSMA-PET positive high-risk mCRPC progressed on androgen receptor signaling inhibitor such as enzalutamide or abiraterone, using 3 mL of blood samples, CTC enumeration and PSMA protein expression were demonstrated with the PSMA CTC assay developed in cancer CLCs. Staining, classification of the slides, and analysis of the 24 mCRPC samples revealed that 21/24 (88%) had at least one traditional CTC [CK(+), CD45(−), and intact DAPI] per milliliter of blood (ranging from 0 to 30 CTCs/mL across the cohort). Of these, 76% (16/21) cases had PSMA protein expression positive CTCs (CK+, CD45−, and DAPI), ranging from 0 to 27 CTCs/mL of blood, suggesting there might be tumor heterogeneity (PSMA-positive and PSMA-negative CTCs; Fig. 3A). Example CTCs in mCRPC patients with PSMA staining are shown in Figure 3B.
Low-pass whole-genome sequencing at single CTC level
To identify the CNV, 36 single CTCs from 11 patients were picked, amplified, and low-pass whole-genome sequenced. The sequencing data was analyzed using a copy number analysis pipeline, where read counts were binned and corrected for known biases. The resulting bin counts were used for CNV calling. The CTCs from 11 patients that underwent low-pass whole-genome sequencing revealed genomic heterogeneity; frequent genomic amplification of the genes AR (27%), MYC (64%), and MYCN (45%); and copy loss of PTEN (36%), TP53 (64%), BRCA2 (45%), CHD1 (55%), and RB1 (45%) (Fig. 4A). Genomic variation at the chromosomal level and at the single CTC level is shown in Figure 4B.
Discussion and conclusion
In this article, we detailed the development of a PSMA blood-based CTC assay utilizing Epic Sciences’ PSMA CTC assay platform. We performed analytical validation on LD slides and clinical validation on CTC samples from men with mCRPC with documented PSMA expression on PET scans. The analytical validation demonstrated high performance, which firmly supported its capability to identify the PSMA protein expression in CTCs from high-risk patients with mCRPC. Further, Epic Sciences’ PSMA CTC assay platform can be coupled with single-cell CTC sequencing to evaluate CNV and genomic heterogeneity in late-stage disease.
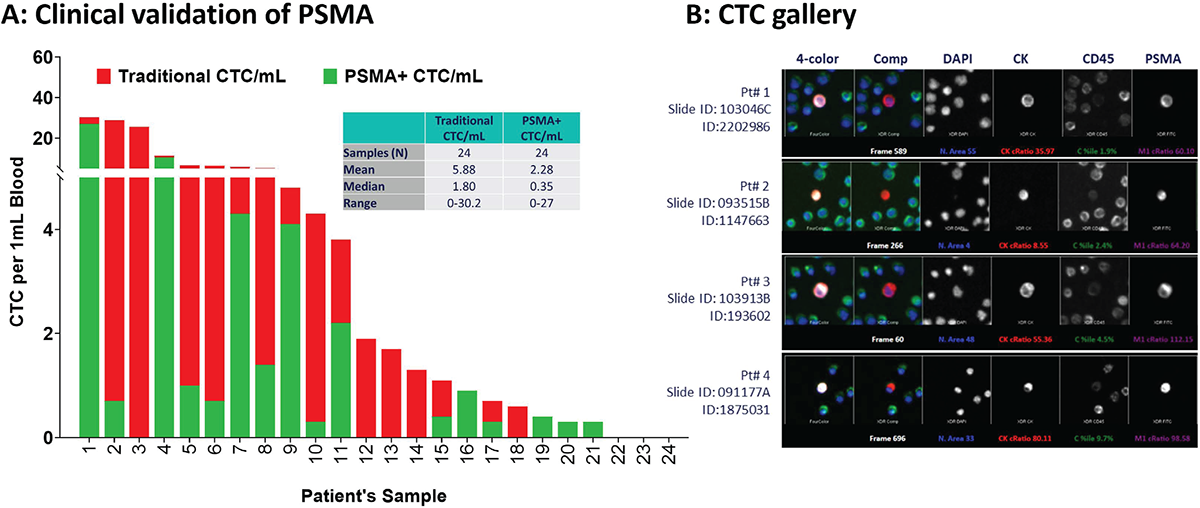
FIGURE 3 - Clinical validation of PSMA protein expression in CTCs of men with mCRPC. A) Analysis of 24 mCRPC samples revealed that 21/24 (88%) had at least one conventional CTC/mL (range: 0-30 CTCs/mL), of which 16/21 (76%) were PSMA positive (range: 0-27 CTCs/mL) with considerable tumor heterogeneity. B) A representative gallery of CTCs with PSMA expression in four unique CTCs in mCRPC patients. CTC = circulating tumor cell; mCRPC = metastatic castration-resistant prostate cancer; PSMA = prostate-specific membrane antigen.
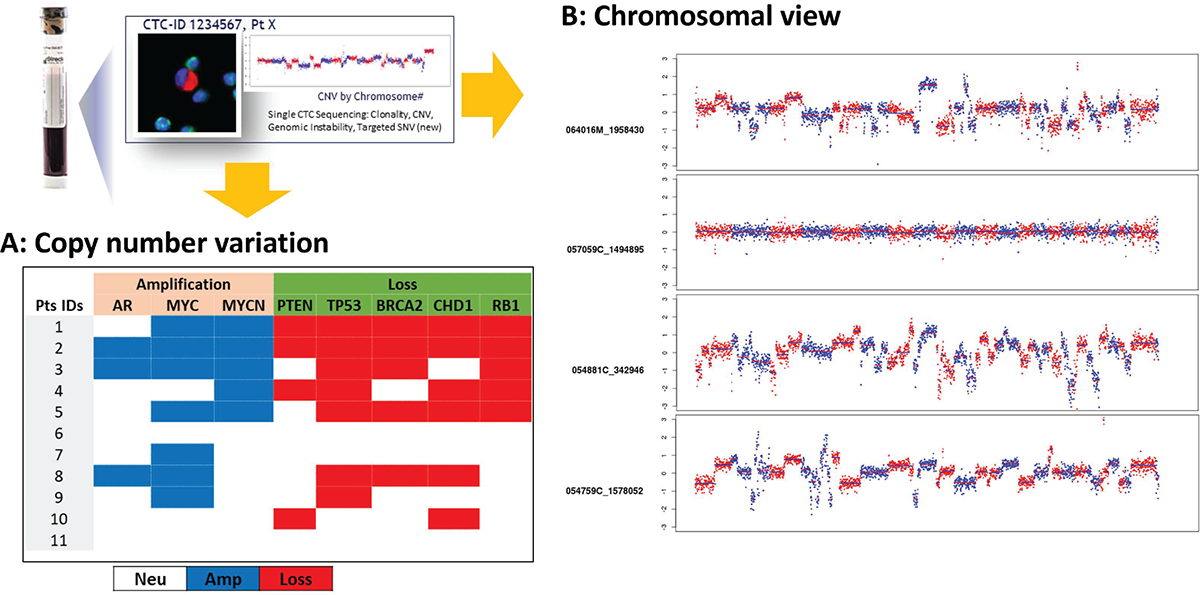
FIGURE 4 - Individual CTCs can be identified and collected for next-generation sequencing to identify CNVs. A) 36 CTCs sequenced from 11 patients exhibited genomic heterogeneity and frequent AR (3/11), MYC (7/11), and MYCN (5/11) amplification and loss of PTEN (4/11), TP53 (7/11), BRCA2 (5/11), CHD1 (6/11), and RB1 (5/11). B) Genome-wide chromosomal view images showing CNV at the single CTC level in mCRPC patients. CNV = copy number variation; CTC = circulating tumor cell; mCRPC = metastatic castration-resistant prostate cancer.
Expression of PSMA in mCRPC makes PSMA an attractive drug target. PSMA-targeted therapies are used in the management of metastatic prostate cancer and may have potential for wider usage in personalized management of metastatic prostate cancer (4). The PSMA protein, which is present on the plasma membrane of cells, is overexpressed in over 90% of individuals with mCRPC, and independently linked to shorter PFS and OS (4). Novartis recently received FDA approval for the PSMA RLT, 177Lu-PSMA-617, based on the improvement in PFS and OS from 177Lu-PSMA-617 in conjunction with best SoC compared with SoC alone in men with mCRPC who had previously received potent androgen receptor pathway inhibitors and chemotherapy in the phase III VISION trial (4). In numerous research studies, PET imaging has been utilized to validate that patients have a curable PSMA-specific condition and track response to treatment. When compared to traditional imaging (CT, MRI), PSMA-PET is far more sensitive and specific to PSMA for detecting micro-metastatic illness at both initial staging and biochemical recurrence (NCCN 2021). Although PSMA-negative tumor cells might not be visible by PSMA-PET imaging, they might nevertheless contribute to the development of progressive disease, particularly in heavily treated mCRPC. The correlation between PSMA-PET imaging parameters and PSMA-positive and PSMA-negative CTC enumeration is an area of active investigation. Additionally, serial CTC profiling to monitor response to PSMA-targeted therapy (traditionally based on PSA and conventional imaging and/or PET) and associations with clinical outcomes require further research and validation in large trial datasets. This research effort could help to identify the 25%-30% of men with mCRPC who exhibit primary resistance to PSMA RLT, thereby mitigating additional exposure and allowing early transition to alternate therapies (17). The analytical results of the Epic Sciences’ PSMA CTC assay clearly confirm its viability for detecting PSMA expression and for single-cell whole-genome sequencing in CTCs from patients with mCRPC to identify genomic abnormalities known to be recurrent in this disease phase (AR, PTEN, TP53, CHD1, and BRCA1/2). High levels of sensitivity, specificity, and overall accuracy were shown by the PSMA CTC test.
Finally, we discussed the development of a PSMA blood-based assay for CTC identification and the implementation of the low-pass whole-genome single-cell sequencing workflow to detect variations in copy number at the genome level and to further define CTC tumor heterogeneity in late-stage high-risk mCRPC. This test is positioned as a potent research tool to detect and prospectively monitor PSMA-positive CTC expression and examine tumor heterogeneity, therapeutic response, and resistance in these mCRPC patients. The PSMA-avid disease is the focus of numerous ongoing clinical trials, including PRINCE (NCT03658447) and the LuPARP study (NCT03874884), which utilize this powerful assay for early disease identification, disease monitoring, and understanding drug resistance in patients with mCRPC.
Abbreviations: CLC = cell line cell; CNV = copy number variation; CT = computerized tomography; CTC = circulating tumor cell; DNA = deoxyribonucleic acid; FDA = Food and Drug Administration; HD = healthy donor; ICC = intra-class correlation; IF = immunofluorescence; LD = lab derived; LOD = limit of detection; mCRPC = metastatic castration-resistant prostate cancer; MRI = magnetic resonance imaging; NPV = negative predictive value; OS = overall survival; PET = positron emission tomography; PFS = progression-free survival; PPV = positive predictive value; PSA = prostate-specific antigen; PSMA = prostate-specific membrane antigen; RLT = radioligand therapy; SoC = standard of care; TSA = tyramide signal amplification; WBC = white blood cell
Disclosures
Conflict of Interest: The authors declare no conflict of interest.
Financial support: This research was supported by Epic Sciences, Victorian Cancer Agency, Australia and Prostate Cancer Foundation.
References
- 1. Bakht MK, Derecichei I, Li Y, et al. Neuroendocrine differentiation of prostate cancer leads to PSMA suppression. Endocr Relat Cancer. 2018;26(2):131-146. CrossRef PubMed
- 2. Kaittanis C, Andreou C, Hieronymus H, et al. Prostate-specific membrane antigen cleavage of vitamin B9 stimulates oncogenic signaling through metabotropic glutamate receptors. J Exp Med. 2018;215(1):159-175. CrossRef PubMed
- 3. Zaman MU, Fatima N, Zaman A, Sajid M, Zaman U, Zaman S. Diagnostic challenges in prostate cancer and 68Ga-PSMA PET imaging: a game changer? Asian Pac J Cancer Prev. 2017;18(10):2625-2628. PubMed
- 4. Sartor O, de Bono J, Chi KN, et al; VISION Investigators. Lutetium-177-PSMA-617 for metastatic castration-resistant prostate cancer. N Engl J Med. 2021;385(12):1091-1103. CrossRef PubMed
- 5. Lapidus RG, Tiffany CW, Isaacs JT, Slusher BS. Prostate-specific membrane antigen (PSMA) enzyme activity is elevated in prostate cancer cells. Prostate. 2000;45(4):350-354. CrossRef PubMed
- 6. Ashworth TR. A case of cancer in which cells similar to those in the tumors were seen in the blood after death. Australas Med J. 1869;14:146-149.
- 7. Armstrong AJ, Luo J, Nanus DM, et al. Prospective multicenter study of circulating tumor cell AR-V7 and taxane versus hormonal treatment outcomes in metastatic castration-resistant prostate cancer. JCO Precis Oncol. 2020;4:PO.20.00200. CrossRef PubMed
- 8. Deng Z, Wu S, Wang Y, Shi D. Circulating tumor cell isolation for cancer diagnosis and prognosis. EBioMedicine. 2022;83:104237. CrossRef PubMed
- 9. Alix-Panabières C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer. 2014;14(9):623-631. CrossRef PubMed
- 10. Cohen SJ, Punt CJ, Iannotti N, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26(19):3213-3221. CrossRef PubMed
- 11. de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14(19):6302-6309. CrossRef PubMed
- 12. Boudadi K, Suzman DL, Anagnostou V, et al. Ipilimumab plus nivolumab and DNA-repair defects in AR-V7-expressing metastatic prostate cancer. Oncotarget. 2018;9(47):28561-28571. CrossRef PubMed
- 13. De Bono JS, Pantel K, Efstathiou E, et al. CTC counts as a biomarker of prognosis and response in metastatic castration-resistant prostate cancer (mCRPC) from the CARD trial. J Clin Oncol. 2021;39(6_suppl):161. CrossRef
- 14. Armstrong AJ, Halabi S, Luo J, et al. Prospective multicenter validation of androgen receptor splice variant 7 and hormone therapy resistance in high-risk castration-resistant prostate cancer: the PROPHECY Study. J Clin Oncol. 2019;37(13):1120-1129. CrossRef PubMed
- 15. Greene SB, Dago AE, Leitz LJ, et al. Chromosomal instability estimation based on next generation sequencing and single cell genome wide copy number variation analysis. PLoS One. 2016;11(11):e0165089. CrossRef PubMed
- 16. Armstrong AJ, Gupta S, Healy P, et al. Pharmacodynamic study of radium-223 in men with bone metastatic castration resistant prostate cancer. PLoS One. 2019;14(5):e0216934. CrossRef PubMed
- 17. Hamid A, Hofman MS, Bressel M, et al. Circulating tumour cells (CTCs) and PSMA PET correlates in the phase I PRINCE trial of 177Lu-PSMA-617 plus pembrolizumab for metastatic castration resistant prostate cancer (mCRPC). J Clin Oncol 2022;40(16_suppl):5027. CrossRef
- 18. Lu D, Krupa R, Harvey M, et al. Development of an immunofluorescent AR-V7 circulating tumor cell assay – A blood-based test for men with metastatic prostate cancer. J Circ Biomark. 2020 Oct 23;9:13-19. PubMed