 |
Drug Target Insights 2025; 19: 18-30 ISSN 1177-3928 | DOI: 10.33393/dti.2025.3368 ORIGINAL RESEARCH ARTICLE |
 |
Withania somnifera root extract (LongeFera™) confers beneficial effects on health and lifespan of the model worm Caenorhabditis elegans
ABSTRACT
Background: Withania somnifera is among the most widely prescribed medicinal plants in traditional Indian medicine. Hydroalcoholic extract of the roots of this plant was investigated for its effects on the overall health and lifespan of the model worm Caenorhabditis elegans.
Methods: The extract’s effect on worm lifespan and fertility was observed microscopically. Worm motility was quantified through an automated worm tracker. The metabolic activity of the worms was captured using the Alamar Blue® assay. Differential gene expression in extract-treated worms was revealed through a whole transcriptome approach.
Results: Extract-exposed gnotobiotic worms, in the absence of any bacterial food, registered longer lifespan, higher fertility, better motility, and metabolic activity. Whole transcriptome analysis of the extract-treated worms revealed the differential expression of the genes associated with lifespan extension, eggshell assembly and integrity, progeny formation, yolk lipoproteins, collagen synthesis, cuticle molting, etc. This extract seems to exert its beneficial effect on C. elegans partly by triggering the remodeling of the developmentally programmed apical extracellular matrix (aECM). Differential expression of certain important genes (cpg-2, cpg-3, sqt-1, dpy-4, dpy-13, and col-17) was confirmed through PCR assay too. Some of the differently expressed genes (gfat-2, unc-68, dpy-4, dpy-13, col-109, col-169, and rmd-1) in worms experiencing pro-health effect of the extract were found through co-occurrence analysis to have their homologous counterpart in humans.
Conclusions: Our results validate the suitability of W. somnifera extract as a nutraceutical for healthy aging.
Keywords: Caenorhabditis elegans, Fertility, Healthy aging, Healthspan, Longevity, Network analysis, Nutraceutical, Withania somnifera root, Worm transcriptome
Received: November 4, 2024
Accepted: March 5, 2025
Published online: April 2, 2025
This article includes supplementary materials
Corresponding author:
Vijay Kothari
email: vijay.kothari@nirmauni.ac.in
Drug Target Insights - ISSN 1177-3928 - www.aboutscience.eu/dti
© 2025 The Authors. This article is published by AboutScience and licensed under Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0).
Commercial use is not permitted and is subject to Publisher’s permissions. Full information is available at www.aboutscience.eu
Introduction
For centuries, humans have longed for a long and healthy life. A variety of plant extracts and polyherbal formulations have been claimed to impart the same in different systems of traditional medicine. However, the widespread application of such plant products in modern times requires scientific validation of the claimed activities. One of the most popular plants in the Indian system of Ayurved is Withania somnifera (L.) Dunal, commonly known as Ashwagandha. This plant belongs to the Solanaceae family and has a long history of >3000 years of being prescribed in indigenous medicine (1). In Ayurved, W. somnifera has been referred to as ‘Rasayan,’ which means a holistic therapy for suppressing the aging process, developing positive physical and mental health, boosting the immune system, and maintaining youth. Ancient texts mention W. somnifera as ‘Avarada,’ the meaning of which is related to longevity or regeneration. The multitude of activities (2) claimed in this plant include immunomodulatory, anti-inflammatory, neuroprotection, anticancer, anti-stress, and pro-fertility (3). Its regular consumption is believed to rejuvenate the reproductive organs and promote fertility, retard senescence, and relieve nervous exhaustion, sexual debility, muscular weakness, and geriatric problems. W. somnifera is expected to inhibit aging and catalyze the anabolic processes of the body (4). Among all parts of this plant, its root has received maximum attention in medicinal texts and practice. This is because its roots have the highest concentration of desired bioactive principles (5). Further, with respect to safety, only the root part of W. somnifera is recommended for therapeutic and internal administration (6).
Mainstreaming the use of any herbal preparation for therapeutic as well as nutraceutical applications is dependent on the demonstration of claimed biological activities in them through appropriate scientific assays (7). Owing to concerns of ethics and feasibility, assays with higher animals and human volunteers are often challenging. Lower animals like the nematode worm Caenorhabditis elegans offer a useful and relatively convenient and biologically relevant platform for assessment of biological activities in natural as well as synthetic preparations at the whole organism level. In recent years, wild-type and transgenic strains of C. elegans have been employed widely as model organisms for assays (8) relevant to neurology, lifespan, diabetes, wound healing (9), and microbial virulence (10). Aging is a complex and multifactorial phenomenon, and it is difficult to model it, and more so, the assessment of longevity or lifespan within a shorter time frame. Though C. elegans has a short lifespan (a few days), the fundamental biological mechanisms and systems in this worm are similar to the mammalian system (11). The mechanisms associated with the increase in longevity identified in this worm are shown to follow a pattern similar to those in humans (12,13). Though the lifespan-enhancing effect of W. somnifera root extract has been reported earlier, too (14), much remains to be elucidated with respect to the underlying molecular mechanisms. This study attempted to fill the said gap by investigating the effect of the hydroalcoholic extract of W. somnifera root on the lifespan and overall healthspan of C. elegans through in vivo assays as well as gene expression analysis at the whole transcriptome level.
Methods
Plant extract
The hydroalcoholic extract (LongeFera™) of Withania somnifera root was procured from Phytoveda Pvt. Ltd., Mumbai. The roots of W. somnifera were collected from Madhya Pradesh, India. It was authenticated by a taxonomist at the Botanical Survey of India, Jodhpur, and the voucher specimen was deposited there with reference ID: BSI/AZRC/I.12012/Tech/19-20/PI.Id/671. The root material was washed, dried, and then pulverized. The resulting powder was then extracted with ethanol: water (8:2 v/v) at 60°C. A powdered form of this extract was then used for assay purposes. The extract was analyzed as per United States Pharmacopeia (USP) (15) for various quality control parameters. The content of withanosides and withanolides was determined using HPLC. Major phytocompounds identified in the extract are depicted in the supplementary chromatogram (Figure S1). The total withanolides content in the extract was found to be 2.69 ± 0.02%.
The extract powder was divided into two batches. One was suspended in water, and another in DMSO (Merck) for bioassay purposes. While the extract was fully soluble in DMSO, its insoluble fraction from the aqueous suspension was removed by centrifugation (7500 g at 25°C) for 10 minutes. The soluble fraction (supernatant) was filtered through a syringe filter (0.45 µm; Axiva), and the filtrate was stored in a sterile 15 mL glass vial (Borosil) under refrigeration. The solubility of the extract in water was 81.84%.
Test organism
Wild type N2 Bristol strain of C. elegans procured from the Caenorhabditis Genetics Center (Minneapolis, USA) was used as a model organism in this study. Lyophilized E. coli OP50 procured from Biovirid (Netherlands) was used as food for C. elegans while maintaining the worm on NGM agar plates (Nematode Growing Medium). Worm synchronization was done as described in literature (16). Prior to the in vivo assays, worms were kept without food for two days to make them gnotobiotic.
Lifespan assay
Synchronized (L3-L4 stage) gnotobiotic worms were incubated at 22 ± 1°C with different concentrations (5-1000 µg/mL) of the plant extract in 24-well plates (HiMedia). Each well contained 1 mL total volume (995 µL M9 buffer + 5 µL extract). Control wells contained only M9 buffer with worms but no extract. Ten worms were added in each well, and these 24-well plates were monitored over a 12-day period (till all worms died in the control wells) under a microscope (4× objective) for live-dead counting. Appropriate vehicle control containing 0.5%v/v DMSO was also included in the experiment.
Motility assay
Synchronized worms were incubated at 22±1°C with or without extract (600 ppm) in 24-well plates, wherein each well contained approximately 100 worms in 1 mL of M9 buffer. While microscopic observation qualitatively confirmed more agile movement in worm population incubated with extract, quantification of the motility in control vs. experimental wells was achieved employing an automated worm tracker machine, WMicrotracker ARENA (Phylumtech, Argentina). Worm activity counts were recorded once every day till progenies appeared in the extract-containing wells (because the presence of progenies will contribute to higher activity counts).
Metabolic activity assay
Alamar Blue® assay (17,18) was used to quantify the viability or metabolic activity of the worms. Synchronized worms were incubated at 22 ± 1°C with or without extract (600 ppm) in 24-well plates, wherein each well contained approximately 100 worms in M9 buffer. One hundred µL of Alamar Blue® (Thermofisher) was added into each well, making the total volume 1 mL, before incubation started. To quantify the amount of dye reduced, after every 24 h, content from wells was transferred into a separate plastic vial (1.5 mL), followed by centrifugation (13,600 g at 25°C) for 10 min. Then, the supernatant was read at 570 nm (Agilent Cary 60 UV-vis). A total of three wells were set for the control as well as the experimental worm population, and content from one well from the control and experimental group each was used on a particular day. Appropriate abiotic controls (containing the dye and other media components but no worms) were also included in the assay.
Whole Transcriptome Analysis
To unravel the molecular mechanisms through which W. somnifera conferred beneficial effects on the worms with respect to lifespan and fertility, the gene expression pattern of extract-treated worms was compared with that of the control worm population at the whole transcriptome scale. While harvesting the worms for RNA isolation on the seventh day of the experiment (when ~50% of worms in the control population were dead), progenies were separated from the experimental population by resting the tubes containing worms incubated with extract in static condition for a few minutes, allowing settling down of adult worms. Progenies were removed by collecting the upper layer of liquid. This removed liquid was microscopically observed to confirm the presence of largely progeny worms in it, and not the adult ones added at the starting day of the experiment. Following this, worms were washed thrice with sterile M9 buffer before proceeding further. The overall workflow of this whole transcriptome analysis (WTA) aimed at capturing a holistic picture regarding modes of action of the test extract is given in Figure S2, along with full methodological details.
All the raw sequence data were submitted to the Sequence Read Archive. The relevant accession number for the control and experimental worm population is SRX20790765 (https://www.ncbi.nlm.nih.gov/sra/SRX20790765) and SRX20790719 (https://www.ncbi.nlm.nih.gov/sra/SRX20790719) respectively.
Polymerase Chain Reaction (RT-PCR)
Differential expression of the potential hubs found using network analysis of DEG revealed from WTA was further confirmed using PCR. Primer designing for the shortlisted genes was carried out using Primer3 Plus. These primer sequences (Table 1; Figure S3) were checked for their binding exclusivity to the target gene sequence within the whole C. elegans genome. Following RNA extraction, cDNA synthesis was carried out using the synthesis kit SuperScript™ VILO™ (Invitrogen Biosciences). PCR assay employed the gene-specific primers purchased from Sigma-Aldrich. FastStart Essential DNA Green Master mix (Roche, Germany) was used as the reaction mix. The real-time PCR assay was performed on a Quant Studio 5 real-time PCR machine (Thermo Fisher Scientific, USA). The temperature profile followed is given in Table S2.
Statistics
All values reported are derived from three or more independent experiments, wherein each experiment contained three replicates (unless specified otherwise). Statistical significance was assessed using a t-test performed in Microsoft Excel® (Version 2016), and data with p ≤ 0.05 were considered to be significant.
Results and Discussion
Worms incubated with the root extract exhibited extended lifespans and higher fertility rates than the control population
We incubated worms with the water-soluble fraction of the root extract, or that dissolved in DMSO, and observed the worms over a period of 12 days (till almost all worms in control wells died) for live-dead count, morphology, agility, and whether any progenies were formed. While the comparison between control and experimental wells was made on a daily basis, two time points can particularly be considered important, i.e., the days on which the control population exhibited ~50% and ~100% death. With respect to these two endpoints, while the DMSO-dissolved extract could impart a longevity benefit to the worms at all tested concentrations ≥100 µg/mL, the water-soluble fraction of the extract could do so at ≥250 µg/mL. Additionally, concentrations ≥ 500 µg/mL of both DMSO-solubilized and water-solubilized extract supported worm fertility from day 4 onward, as evident from the appearance of progenies in the extract-containing wells and their absence in control wells (Videos S1–S2). Till day 12, these progenies could be differentiated from the parent worms based on size. Thereafter, almost all the parent worms in control wells were dead, and the size of the surviving parent worms and their progenies in the experimental wells became similar to the extent that they could not be differentiated. The magnitude of survival benefit (i.e., the higher number of surviving worms in extract-containing wells than in control wells) conferred on the worms by DMSO-solubilized extract (Figure 1A) was somewhat higher than that of water-solubilized extract (Figure 1B), except at 750 µg/mL. At the latter concentration, the water-solubilized fraction of the extract performed better (p = 0.0004) than the DMSO-solubilized extract, as per the final-day endpoint. We decided to perform further experiments with water-solubilized fraction only, as water-solubility of any therapeutic preparation is looked at favorably with respect to bioavailability (19).
| Gene ID/ Name | Primers | Amplicon size (bp) |
|---|---|---|
|
WBGene00005016 (sqt-1) |
FP:
5’-GTTCCAGGACTTGACGGTGT -3’ |
234 |
| RP:
5’-TCCGATCTTTCCGATTTGAC -3’ |
||
|
WBGene00001066 (dpy-4) |
FP:
5’- ATCACCCTCCCAATGGTGTA -3’ |
172 |
| RP:
5’- CGCATTGCTCGTTGTAGGTA -3’ |
||
|
WBGene00000606 (col-17) |
FP:
5’- AACTGAGAGCCGTGAGAAGC -3’ |
156 |
| RP:
5’- TGATCATTTGGAGCATCTGG -3’ |
||
|
WBGene00001074 (dpy-13) |
FP:
5’- TGCTTAGCCATGGACATTGA -3’ |
227 |
| RP:
5’- TGCAGGTAAGGGCTTCGTTA -3’ |
||
|
WBGene00011063 (cpg-3) |
FP:
5’-TCGGAATCCTCTCGAACATC -3’ |
172 |
| RP:
5’-GCTCCAATGCATTTTCCACT -3’ |
||
|
WBGene00015102 (cpg-2) |
FP:
5’- CCATCCAAATGGAGTTTGCT -3’ |
223 |
| RP:
5’- AGTGCAAGCAGTGAATGACG -3’ |
||
|
WBGene00014018 (Endogenous control) |
FP:
5’- AGCGGAAAGATTTCAGACGA-3’ |
187 |
| RP:
5’- CGATAAATGTGCTCCGGAAT-3’ |
While the results described in Figure1A-1B demonstrated the beneficial effect of test extract on worm lifespan as well as fertility, to investigate its effect on worm longevity separately from fertility, we repeated this assay using FUdR (5-fluoro-20-deoxyuridine; HiMedia)- pre-treated worms, wherein eggs were allowed to hatch on FUdR-containing plates pre-seeded with E. coli OP50, and then resulting L3-L4 stage worms were washed with M9 buffer before being transferred to a fresh NGM agar plate for further use. FUdR can sterilize the adult nematode worms without affecting their development (20). However, FUdR-pre-treated worms could not benefit from the pro-longevity effect of the water-soluble fraction of the W. somnifera root extract except at 750 µg/mL on day 12 (Figure 1C), and here, too, the effect was much lesser than when FUdR was not used (Figure 1B). These results raise caution regarding the use of FUdR in lifespan assays with C. elegans, as this may lead to false-negative conclusions. Though the practice of using FUdR-treated worms is popular (in order to avoid the laborious separation of offspring from adults during the reproductive period) among the worm researchers studying longevity and aging, our results show that FUdR can prevent the detection of pro-longevity activity in a bioactive extract, even when the said activity is there. This corroborates with the earlier suggestion by Aitlhadj and Stürzenbaum (21) that the effect of FUdR is not neutral and owing to its mechanism of actions (inhibition of DNA and RNA synthesis, death of mitotic cells, and the inhibition of protein synthesis), its inclusion in the assays may result in misinterpretation. Hence, it can be suggested that while investigating any natural product’s biological effect in the C. elegans model, it is more useful to do a wholistic assay assessing multiple parameters like lifespan, health, metabolic activity, fertility, etc., rather than doing an assay exclusively focusing on longevity using additional chemicals like FUdR which may prove to be a confounding factor.
W. somnifera root extract positively affects worm motility and metabolic activity
Worms incubated in the presence of the test extract exhibited more active movement (Figure 2A), which can be considered another indicator of overall good health. This observation matches well with the high fertility of extract-exposed worms described in the preceding section since the egg-laying active state and the defecation motor program are both linked to changes in forward and reverse locomotion (22,23). Automated monitoring of C. elegans’ movement is a useful and faster healthspan-based method to study aging (24). Besides positively impacting worm lifespan, fertility, and motility, the root extract also had a stimulatory effect on worm metabolic activity (Fig. 2B). Worms incubated in the presence of the extract were found to have higher reducing potential (captured in terms of their ability to reduce the dye Alamar blue), which can be taken as an indication of better health, as healthy living cells maintain a reducing state within their cytosol (25).
Overall, the root extract imparted its beneficial effect on multiple health parameters of the worm, i.e., lifespan, fertility, motility, and metabolic activity. Although many natural products have been reported to impart pro-longevity effects on different biological model organisms based on lifespan assays, which is a conventional method to monitor aging, it should be noted that a compound or formulation that extends lifespan may not necessarily maintain health (26). Lifespan extension makes sense only if it does not cost on the healthspan front (27). Increased egg laying in extract-exposed older adults reflects an increase in the number of eggs expelled per vulval opening, as well as longer active behavior states. Since the timing of expulsive behaviors (defecation and egg laying) is regulated by sensory mechanisms that detect changes in internal pressure and/or stretch to maintain homeostasis (28), we believe that the test extract had a multifactorial impact on worm physiology at different levels. Feedback of successful egg laying in the presence of the extract might have signaled the germ line to continue the production of oocytes for fertilization. A potential trade-off between reproductive capacity and somatic maintenance in C. elegans has already been mentioned in earlier published literature. Higher levels of progeny production are a biomarker for studying aging and correlate positively with a longer lifespan (29,30). Prolonged reproduction can have a beneficial impact on lifespan.
Since the most promising effect of the root extract on the worm’s overall health (i.e., lifespan and fertility) was observed while using a water-soluble fraction of the extract, and the maximum beneficial effect was observed at 600-750 µg/mL, we decided to investigate the gene expression pattern of worms exposed to the test extract at 600 µg/mL at the whole transcriptome level. Since the biological effect of both these concentrations (600 and 750 µg/mL) was statistically similar (p>0.05), we went ahead with 600 µg/mL. While all experiments presented in Figure 1-2 were performed with gnotobiotic worms facing starvation, we also compared the effect of this root extract on worms fed with their regular lab food, E. coli OP50. Since the worm’s response to the W. somnifera extract was not affected (data not shown) owing to the presence of the bacterial food, we performed the transcriptome study with gnotobiotic worms only to avoid any possible confounding role of bacteria.
W. somnifera root extract exerts its beneficial effect on the nematode worm by triggering differential expression of multiple genes
A whole transcriptome level comparison of the gene expression profile of the extract-treated worms with their extract-non-exposed counterparts revealed all the DEG in the experimental worm population. Keeping the criteria of log FC ≥1.5 and FDR (false discovery rate) ≤0.05, the differentially expressed gene (DEG) count was 85. However, to have higher confidence in our data interpretation, we set more stringent dual criteria of log FC ≥ 2 and FDR ≤ 0.01 to shortlist the genes for further analysis. A total of 16 upregulated and 29 downregulated DEGs passing the said dual criteria are listed in Table 2. These 45 DEG comprise 0.83% of the total C. elegans genome (~5423 genes). Function-wise categorization of these DEG is presented in Figure 3, and the corresponding heat map (Fig. S4) and volcano plot (Fig. S5) are provided in the supplementary file. A detailed discussion on important up/down-regulated genes and how their differential expression would have contributed to the observed results is provided in the supplementary file “Appendix A.”
Since the transcriptome data has identified multiple genes associated with worm exoskeleton components and muscle structure as differently expressed in extract-fed worms, it is clear that the root extract used in the present study has caused restructuring of the exoskeleton in such a way that age-associated development of stiffness in worm skeleton is delayed, and the worm can age in a healthier fashion. We had conducted transcriptome profiling from the worms after seven days of extract exposure, but the classic signs of old age and fertility loss usually observed in worms at this stage of the life cycle did not yet arise in the presence of the extract. Hence, W. somnifera root extract can be concluded to be capable of delaying aging and senescence. Under routine conditions, at day seven of adulthood, sarcopenia is apparent in histologically examined worms, which leads to behavioral change in terms of reduced motor activity. Reduced motility stems from stiffening and thickening of the cuticle, which itself results from unregulated collagen biosynthesis (31), and the root extract used in this study seems to have affected collagen synthesis in such a way that stiffening/ thickening of the cuticle was delayed in the extract-fed worms, and thereby maintaining them in an active motility state for a longer period.
Identifying the most important DEG through network analysis
To identify the proteins with high network centrality, i.e., hub proteins from among the DEG, we generated a protein-protein interaction (PPI) network of upregulated (Fig. 4A) and downregulated (Fig. 5A) genes separately. We arranged members of the PPI network of upregulated genes in decreasing order of node degree (Table S3), and those with a score of ≥5 were subjected to ranking by different cytoHubba methods. Then, we looked for genes that appeared among the top 5 ranked candidates by ≥6 cytoHubba methods, and five such shortlisted genes (Table 3) were further checked for interactions among themselves, followed by cluster analysis (Fig. 4B). The PPI network generated through STRING showed these five important genes to be distributed among three different local network clusters. Since cpg-3 appeared to be part of two different clusters, and both the cpg genes had multiple edges connecting them together (node degree score was 3 for both of them), we selected these two genes (cpg-2 and cpg-3) for further RT-PCR validation. PCR assay did confirm the upregulation of these two proteins in extract-exposed worms (Fig. 6A).
We arranged members of the PPI network for the downregulated genes (Fig. 5A) in decreasing order of node degree (Table S4), and those with a score of ≥5 were subjected to ranking by different cytoHubba methods. Then, we looked for genes that appeared among the top-6 ranked candidates by ≥6 cytoHubba methods, and six such shortlisted genes (Tabl 4) were further checked for interactions among themselves, followed by cluster analysis (Fig. 5B). Four (sqt-1, dpy-4, dpy-13, and col-17) of these six genes belonged to a single cluster (each with a node degree score of 4) and were chosen for further RT-PCR validation. PCR assay did confirm the downregulation of these proteins in extract-exposed worms (Fig. 6B).
Identifying the most important DEG based on their homology with the human genome
To have an empirical idea about the relevance of the results of this study performed in C. elegans, with respect to W. somnifera’s possible beneficial effect in humans, we conducted a co-occurrence analysis between all the 45 DEG in extract-treated C. elegans and the human genome. This analysis resulted in the identification of 16 genes whose counterparts are present in humans (Fig. 7). The presence of multiple genes in the human genome with homology to those expressed differently in extract-exposed worms indicates a good probability of similar effect on humans as observed in C. elegans in this study. Among the DEG, gfat-2 seemed to have the highest similarity with its homolog in humans. The C. elegans gfat-2 (glutamine-fructose 6-phosphate aminotransferase-2) is 88.3% similar to gfat-1 (glutamine-fructose 6-phosphate aminotransferase-1) in amino acid sequence, and the latter is considered as a longevity gene (32). gfat-1 is the key enzyme of the hexosamine pathway and has been mentioned as a regulator of protein quality control and longevity. Increased functionality of gfat-1 induces endoplasmic reticulum-associated protein degradation and autophagy and correlatively extends lifespan and ameliorates a wide spectrum of proteinopathies. It can be said that W. somnifera root extract promotes health and extends the lifespan of C. elegans through endogenous modulation of protein quality control. The upregulation of gfat-1 in our long-lived worms corroborates with the previously published observation that hexosamine pathway metabolites enhance protein quality control and extend life (33). In mammals, too, gfat is the rate-limiting enzyme of the hexosamine pathway. Discussion on important genes listed in Figure-7 other than gfat-2 has been done in preceding sections.
Conclusion
The present study has demonstrated the efficacy of W. somnifera root extract in extending the lifespan of the model worm C. elegans while simultaneously supporting healthy aging and allowing progeny formation in the absence of any bacterial food. The study found the hydroalcoholic root extract of W. somnifera (LongeFera™) to offer multiple beneficial health effects to the model organism. Worm lifespan and healthspan (motility, metabolic activity, and fertility) were both positively influenced by the test extract. The whole transcriptome analysis of the extract-exposed worms revealed the multiple mechanisms through which the extract would have exerted its pro-health effect (Fig. 8). Among the major modes of action underlying the observed biological effect of this extract was upregulation of the yolk lipoprotein vitellogenins, remodeling of the apical extracellular matrix, modulation of eggshell permeability, alteration of collagen synthesis, cuticle development, and molting cycle. Many of the differentially expressed genes in the extract-fed worms have a homologous counterpart in humans. Considering various parameters like fold-change value, network centrality, cytoHubba ranking, contribution towards more than one function, and homology with the human genome, the most important among all DEG are gfat-2, sqt-1, dpy-13, dpy-4, cpg-3, vit-5, and col-169. The results of this study validate the pro-health efficacy of W. somnifera claimed in traditional medicine systems like Ayurved and strengthen the candidature of this plant as a potential nutraceutical for healthy aging. Our results also raise caution against the use of anti-fertility agents like FUdR in worm assays, as that not only may prove a confounding factor in the interpretation of results but also precludes simultaneous assessment of lifespan, fertility, and other healthspan parameters in a single assay. To the best of our awareness, this is the first report describing molecular mechanisms associated with the beneficial effects of W. somnifera root extract on the model worm C. elegans.
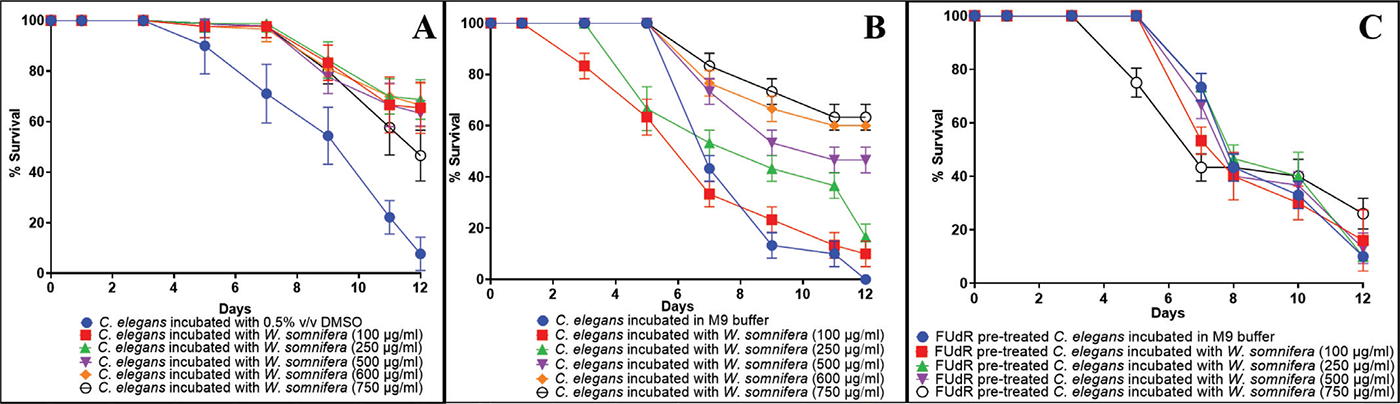
FIGURE 1 - Withania somnifera imparts longevity extension on C. elegans. All worms in control wells (i.e., worms not fed with extract) were dead by the 12th day. Concentrations ≥500 µg/mL of both DMSO-solubilized and water-solubilized extract supported worm fertility from day 4 onward. Bigger body size, higher motility, and progeny formation in the presence of the extract in experimental wells can be visualized in supplementary videos S1–S4. DMSO (0.5%v/v) or FUdR (15 mM/mL of NGM agar) did not affect worm lifespan. Only selected concentrations are displayed in this figure to avoid graph overcrowding. (A) Worms fed with DMSO-solubilized W. somnifera extract at 100, 250, 500, 600, 750 μg/mL scored 65.55%*** ± 10.13, 68.88%*** ± 7.81, 63.33%*** ± 5, 66.66%*** ± 8.66 and 46.66%*** ± 10 better survival on the 12th day compared to control. (B) Worms fed with water-solubilized W. somnifera at 100, 250, 500, 600, 750 μg/mL scored 10%*** ± 5, 16.66%*** ± 5, 46.66%*** ± 5, 60%*** ± 0 and 63.33%*** ± 5 better survival on the 12th day compared to control. (C) FUdR pre-treated worms fed with water-solubilized W. somnifera extract did not display any extended lifespan except at 750 μg/mL (26.66%*** ± 5.16 better survival on the 12th day compared to control. *** p ≤ 0.001
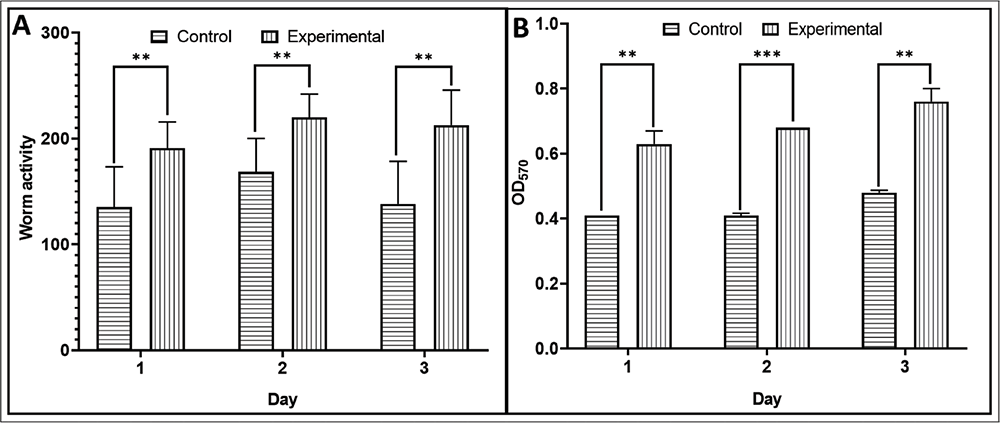
FIGURE 2 - W. somnifera root extract has a positive effect on worm motility (A) and metabolic activity. (B) Worm activity plotted on the y-axis in Figure (A) is the quantification of their movement as measured by an automated worm tracker. Higher motility in extract-treated worms plotted here is in line with the visual observation under the microscope. OD570 in Figure (B) corresponds to the amount of dye reduced by the metabolic activity of the worm population. **p ≤ 0.01, ***p ≤ 0.001; Extract concentration used was 600 µg/mL.
| Sr. No | Gene ID | Gene | Product/function | Up-/Down-regulation | Log fold change |
|---|---|---|---|---|---|
| 1 | WBGene00006929 | vit-5 | Vitellogenin-5 | ↑ | 10.78 |
| 2 | WBGene00012592 | Y38E10A.14 | Uncharacterized protein | ↓ | 9.02 |
| 3 | WBGene00011501 | rmd-1 | Regulator of microtubule dynamics protein 1 | ↑ | 8.06 |
| 4 | WBGene00013391 | Y62H9A.3 | Uncharacterized protein | ↑ | 7.92 |
| 5 | WBGene00019146 | H02F09.3 | Uncharacterized protein | ↓ | 7.75 |
| 6 | WBGene00017501 | pud-3 | PUD domain-containing protein | ↑ | 5.97 |
| 7 | WBGene00012261 | lpr-3 | Lipocalin-Related protein | ↓ | 5.74 |
| 8 | WBGene00021005 | ule-1 | Chitin-binding type-2 domain-containing protein | ↑ | 5.63 |
| 9 | WBGene00012186 | mlt-11 | Uncharacterized protein | ↓ | 5.52 |
| 10 | WBGene00006926 | vit-2 | Vitellogenin-2 | ↑ | 5.36 |
| 11 | WBGene00009429 | irg-5 | CUB like domain-containing protein | ↓ | 5.35 |
| 12 | WBGene00000683 | col-109 | Cuticle collagen N-terminal domain-containing protein | ↓ | 5.04 |
| 13 | WBGene00019148 | H03E18.1 | Uncharacterized protein | ↓ | 5.04 |
| 14 | WBGene00017498 | pud-4 | PUD domain-containing protein | ↑ | 4.97 |
| 15 | WBGene00000742 | col-169 | Cuticle collagen N-terminal domain-containing protein | ↓ | 4.9 |
| 16 | WBGene00022033 | Y65B4BL.1 | Uncharacterized protein | ↓ | 4.9 |
| 17 | WBGene00006930 | vit-6 | Vitellogenin-6 | ↑ | 4.49 |
| 18 | WBGene00000618 | col-41 | Cuticle collagen N-terminal domain-containing protein | ↓ | 4.43 |
| 19 | WBGene00018268 | F41C3.2 | Uncharacterized protein | ↓ | 4.4 |
| 20 | WBGene00016636 | perm-2 | Permeable eggshell | ↑ | 4.35 |
| 21 | WBGene00001074 | dpy-13 | Cuticle collagen dpy-13 | ↓ | 4.27 |
| 22 | WBGene00009035 | gfat-2 | Glutamine-fructose-6-phosphate transaminase (isomerizing) | ↑ | 4.19 |
| 23 | WBGene00017892 | F28B4.3 | Uncharacterized protein | ↓ | 4.16 |
| 24 | WBGene00000606 | col-17 | Collagen | ↓ | 4.04 |
| 25 | WBGene00012468 | Y17G7B.17 | Uncharacterized protein | ↓ | 3.98 |
| 26 | WBGene00022816 | fbn-1 | Fibrillin homolog | ↓ | 3.81 |
| 27 | WBGene00022103 | cdh-12 | Cadherin family | ↓ | 3.81 |
| 28 | WBGene00001066 | dpy-4 | Cuticle collagen N-terminal domain-containing protein | ↓ | 3.78 |
| 29 | WBGene00015102 | cpg-2 | Chondroitin proteoglycan-2 | ↑ | 3.76 |
| 30 | WBGene00010790 | sodh-1 | Alcohol dehydrogenase 1 | ↑ | 3.76 |
| 31 | WBGene00006787 | unc-52 | Basement membrane proteoglycan; Ig-like domain-containing protein | ↓ | 3.7 |
| 32 | WBGene00005018 | sqt-3 | Cuticle collagen 1 | ↓ | 3.58 |
| 33 | WBGene00011063 | cpg-3 | Chondroitin proteoglycan 3 | ↑ | 3.54 |
| 34 | WBGene00007709 | clec-87 | C-type lectin domain-containing protein 87 | ↑ | 3.52 |
| 35 | WBGene00011321 | fil-1 | Fasting Induced Lipase | ↑ | 3.48 |
| 36 | WBGene00007723 | C25F9.2 | DNA-directed DNA polymerase | ↓ | 3.37 |
| 37 | WBGene00005016 | sqt-1 | Cuticle collagen sqt-1 | ↓ | 3.37 |
| 38 | WBGene00010070 | nep-17 | Neprilysin metallopeptidase family | ↓ | 3.28 |
| 39 | WBGene00006436 | ttn-1 | Titin homolog | ↓ | 3.09 |
| 40 | WBGene00006801 | unc-68 | Uncharacterized protein | ↓ | 3.08 |
| 41 | WBGene00002915 | let-805 | Uncharacterized protein | ↓ | 3.07 |
| 42 | WBGene00004130 | ketn-1 | Uncharacterized protein | ↓ | 3.05 |
| 43 | WBGene00000998 | dig-1 | Mesocentin | ↓ | 3 |
| 44 | WBGene00006876 | vab-10 | GAR domain-containing protein | ↓ | 2.89 |
| 45 | WBGene00220238 | ZK185.9 | Uncharacterized protein | ↑ | 2.74 |
Genes in this table are arranged in decreasing order of fold-change value.
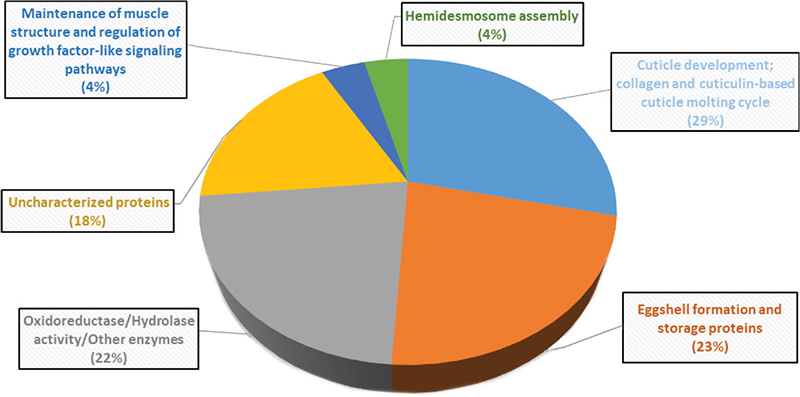
FIGURE 3 - Function-wise categorization of the differentially expressed genes in W. somnifera-exposed C. elegans. Genes contributing to more than one function are considered in any one category only. Values in parentheses are % of the total DEG.
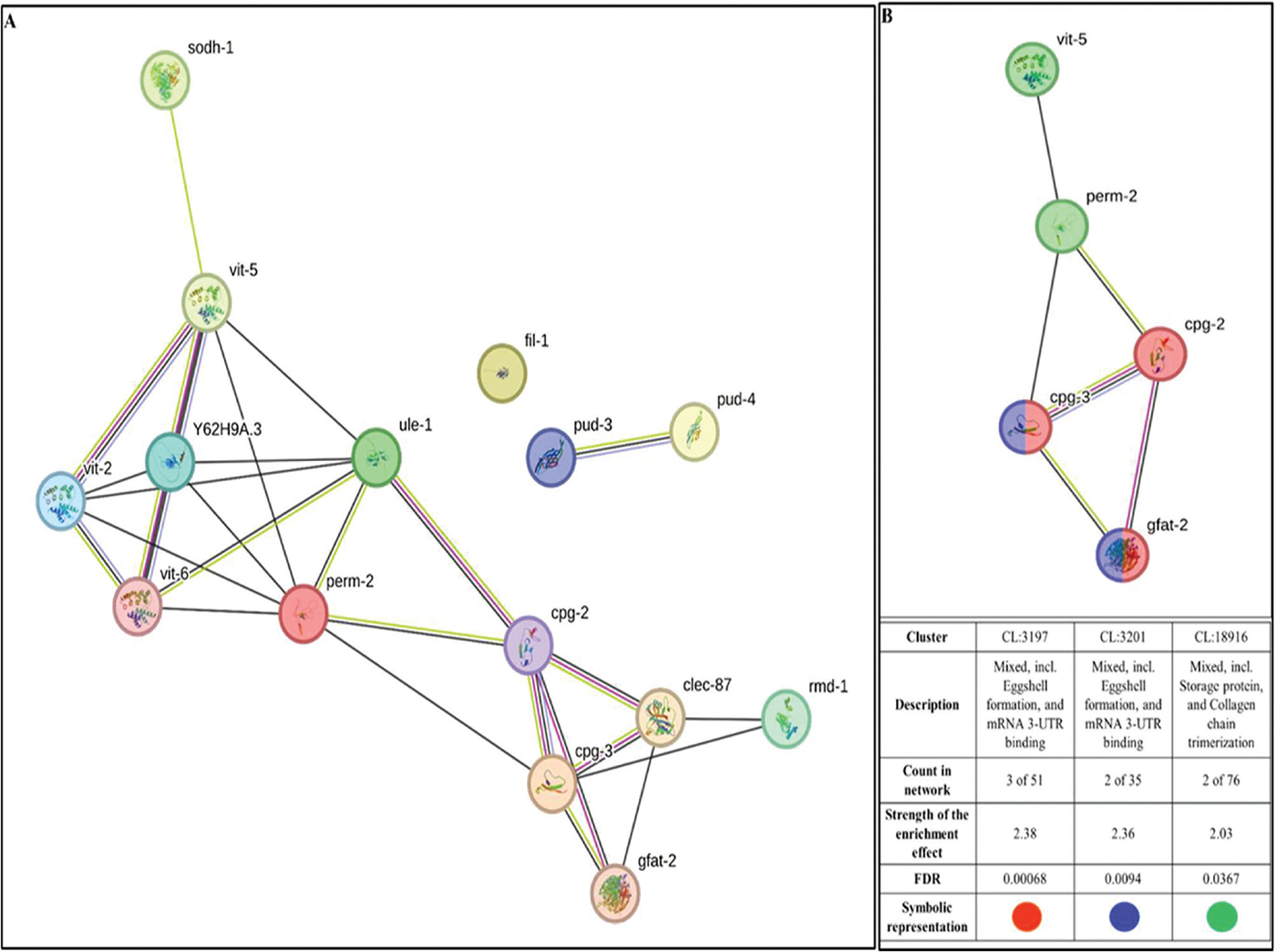
FIGURE 4 - (A) Protein-Protein Interaction (PPI) network of upregulated genes following the dual criteria of fold change ≥ log 2 and FDR ≤ 0.01 in W. somnifera-exposed C. elegans. PPI enrichment p-value 1.0e-16. This network contains 15 nodes connected through 28 edges, with an average node degree of 3.73. Since the number of edges (28) in this PPI network is 28-fold higher than expected (01), this network can be believed to possess significantly more interactions among the member proteins than what can be anticipated for a random set of proteins of the same sample size and degree distribution. (https://string-db.org/cgi/network?taskId=badc9unayQGh&sessionId=bmZFhfliKMsM). (B) PPI network of top-ranked genes shortlisted using cytoHubba among upregulated genes in W. somnifera-exposed C. elegans. PPI enrichment p-value 3.24e-10. With an average node degree score of 2.4, this network possesses more edges (06) than expected (0) for any similar random set of proteins. The value of node degree score for cpg-2, cpg-3, and perm-2 is 3, and that for gfat-2 and vit-5, 2 and 1, respectively. (https://string-db.org/cgi/network?taskId=bbPybwRHQ9La&sessionId=bmZFhfliKMsM)
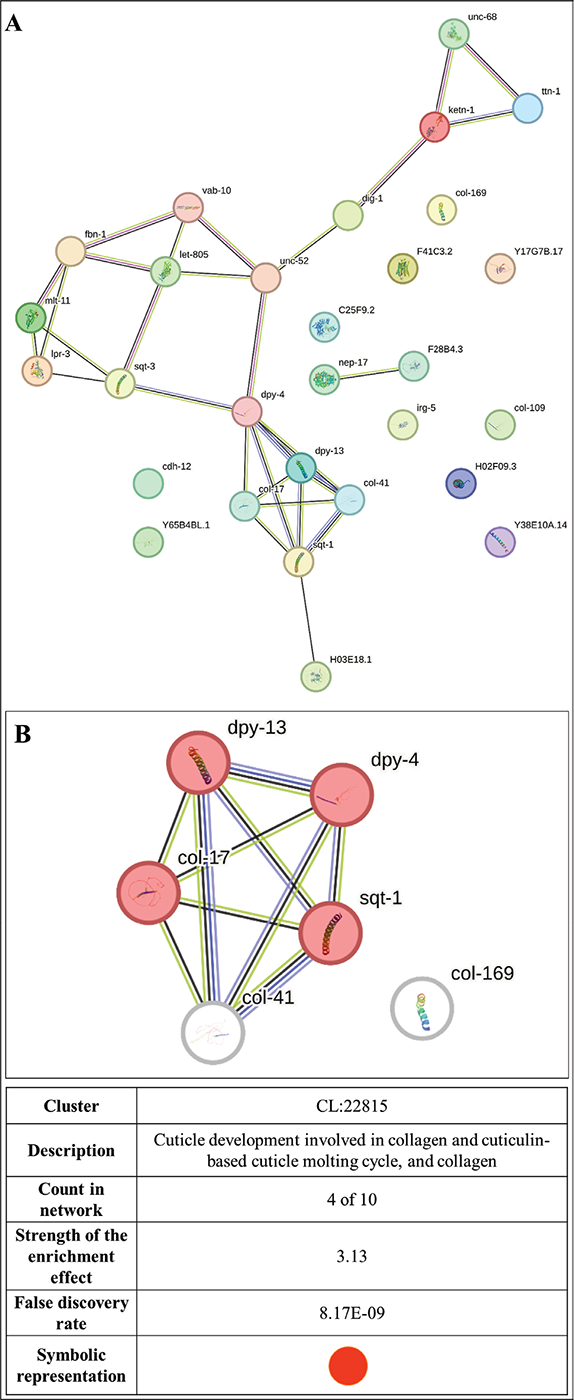
FIGURE 5 - (A) Protein-Protein Interaction (PPI) network of downregulated genes following the dual criteria of fold change ≥ log 2 and FDR ≤ 0.01 in W. somnifera-exposed C. elegans. PPI enrichment p-value 1.0e-16. This network contains 29 nodes connected through 30 edges, with an average node degree of 2.07. Since the number of edges (30) in this PPI network is 30-fold higher than expected (01), this network can be believed to possess significantly more interactions among the member proteins than what can be anticipated for a random set of proteins of the same sample size and degree distribution. (https://string-db.org/cgi/network?taskId=bRhNGHhVNwKu&sessionId=bmZFhfliKMsM). (B) PPI network of top-ranked genes shortlisted using cytoHubba among downregulated genes in W. somnifera-exposed C. elegans. PPI enrichment p-value 1.0e-16. With an average node degree score of 3.88, this network also possessed more edges (10) than expected (0) for any similar random set of proteins. The node degree score of all the genes shown in this network was 4, except col-169 (node degree: zero) (https://string-db.org/cgi/network?taskId=bSBgDIecdxCg&sessionId=bmZFhfliKMsM).
| No. | Gene ID | Gene symbol | No. of methods ranking this protein among the top five | Names of 12 ranking methods of CytoHubba and rank score provided by them | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Degree | MNC | DMNC | MCC | Bottleneck | Eccentricity | Closeness | Radially | Betweenness | Stress | CC | E | ||||
| 1 | WBGene00006929 | vit-5 | 12 | 8 | 8 | 0.758 | 2880 | 1 | 0.5 | 8.5 | 2.222 | 0.571 | 4 | 0.928 | 3.993 |
| 2 | WBGene00016636 | perm-2 | 10 | 9 | 9 | - | 2904 | 2 | 1 | 9 | 2.333 | 3.401 | 14 | – | 4.101 |
| 3 | WBGene00011063 | cpg-3 | 9 | 9 | 9 | - | 2904 | - | 1 | 9 | 2.333 | 3.401 | 14 | – | 4.1 |
| 4 | WBGene00015102 | cpg-2 | 9 | 8 | 8 | - | - | 1 | 0.5 | 8.5 | 2.222 | 2.452 | 10 | – | 3.982 |
| 5 | WBGene00009035 | F22B3.4 | 8 | 8 | 8 | - | - | 1 | 0.5 | 8.5 | 2.222 | 2.452 | 10 | – | – |
‘-’ indicates that the given method did not rank that particular gene among the top five.
| No. | Gene ID | Gene symbol | No. of methods ranking these proteins among the top six | Names of 12 ranking methods of CytoHubba and rank score provided by them | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Degree | MNC | DMNC | MCC | Bottleneck | Eccentricity | Closeness | Radially | Betweenness | Stress | CC | E | ||||
| 1 | WBGene00005016 | sqt-1 | 12 | 5 | 5 | 0.648 | 120 | 1 | 0.5 | 5 | 0.7 | 0 | 0 | 1 | 3.273 |
| 2 | WBGene00001066 | dpy-4 | 12 | 5 | 5 | 0.648 | 120 | 1 | 0.5 | 5 | 0.7 | 0 | 0 | 1 | 3.309 |
| 3 | WBGene00000606 | col-17 | 12 | 5 | 5 | 0.648 | 120 | 1 | 0.5 | 5 | 0.7 | 0 | 0 | 1 | 3.303 |
| 4 | WBGene00000742 | col-169 | 9 | 5 | 5 | 0.648 | 120 | - | 0.5 | 5 | 0.7 | - | - | 1 | 3.243 |
| 5 | WBGene00001074 | dpy-13 | 8 | 5 | 5 | 0.648 | 120 | - | 0.5 | 5 | 0.7 | - | - | - | 3.251 |
| 6 | WBGene00000618 | col-41 | 8 | 5 | 5 | 0.648 | 120 | - | 0.5 | 5 | 0.7 | - | - | - | 3.285 |
‘-’ indicates that the given method did not rank that particular gene among the top six.
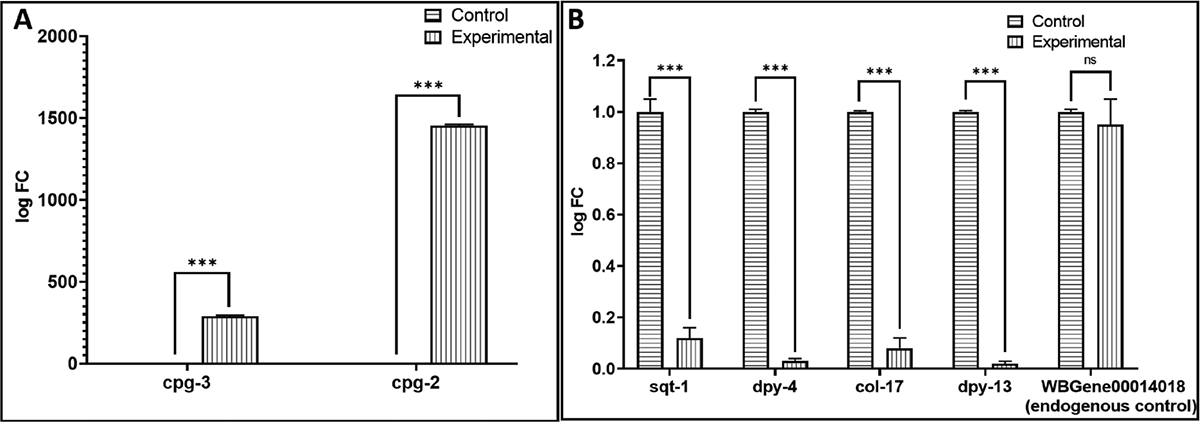
FIGURE 6 - Confirmation of differential expression of selected (A) upregulated and (B) downregulated genes in W. somnifera-exposed C. elegans through RT-PCR.
*** p ≤ 0.001, ns: not significant; The gene employed as endogenous control codes for an RNA-binding protein

FIGURE 7 - Co-occurrence analysis of differentially expressed genes in extract-treated C. elegans with the human genome. The darker the shade of the square, the higher the homology between the genes being compared. Genes from left to right are arranged in descending order of homology. Functions of the genes shown here in humans are listed in Table S5.
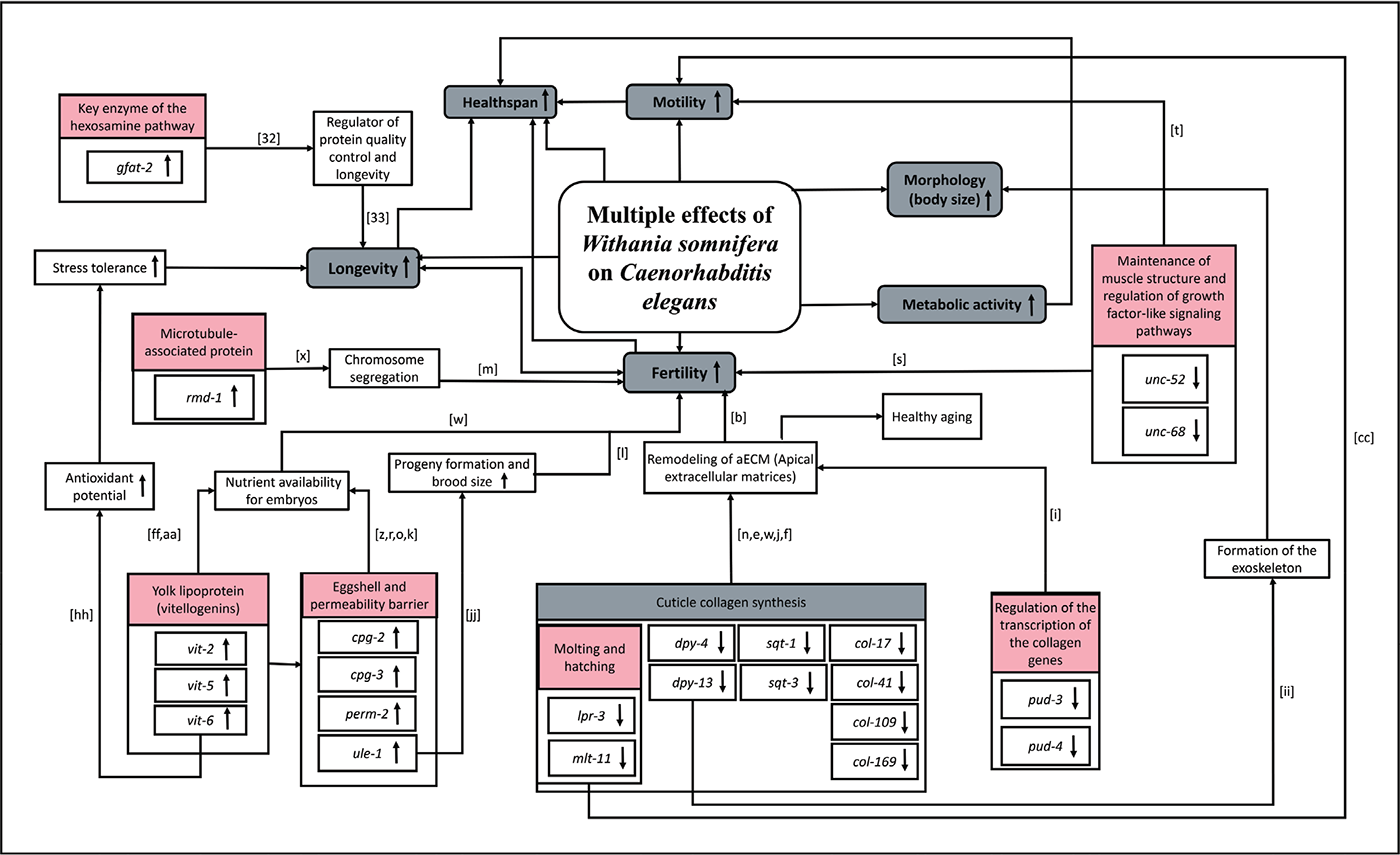
FIGURE 8 - A schematic summary of multiple effects of W. somnifera on C. elegans The small alphabet in square brackets in this figure corresponds to the references cited in the supplementary file “Appendix A.”
Acknowledgements
We thank NERF (Nirma Education and Research Foundation), Ahmedabad, for infrastructural support. This article was initially available as a preprint on BioRxiv on August 2, 2024. CrossRef
Disclosures
Data availability statement: While all other data is provided within the manuscript or supplementary information files, the sequence data pertaining to worm transcriptome have been deposited in the NCBI-SRA database, which can be accessed at: (https://www.ncbi.nlm.nih.gov/sra/SRX20790765) and (https://www.ncbi.nlm.nih.gov/sra/SRX20790719).
Conflict of interest: DM and SN are affiliated with Phytoveda Pvt. Ltd., which markets Withania somnifera extract. However, this in no way has influenced the design of the study or the interpretation of results. NT receives a stipend from NERF; GG acknowledges the scholarship from the Government of Gujarat via their SHODH scheme. The rest of the authors declare no competing interests.
Financial support: This work was funded by Phytoveda Pvt. Ltd.
Author Contributions: Conceptualization: DM and VK; Methodology: GG, NT, and SN; Formal analysis, investigation, data curation: NT, GG, and VK; Resources: DM and VK; Writing—original draft preparation: NT, GG, and VK; Writing—review and editing: NT, GG, SN, DM and VK; Supervision and project administration: VK; Funding acquisition: DM and VK. All authors have read and agreed to the published version of the manuscript.
References
- 1. Bharti VK, Malik JK, Gupta RC. Ashwagandha: multiple health benefits. In: Nutraceuticals 2016 (pp. 717-733). Academic Press. CrossRef
- 2. Kumar S, Mathew SO, Aharwal RP, et al. A pleiotropic anticancer agent from the Indian medicinal plant Withania somnifera (L.) Dunal. Pharmaceuticals (Basel). 2023;16(2):160. CrossRef PubMed
- 3. Sengupta P, Agarwal A, Pogrebetskaya M, et al. Role of Withania somnifera (Ashwagandha) in the management of male infertility. Reprod Biomed Online. 2018;36(3):311-326. CrossRef PubMed
- 4. Baliga MS, Meera S, Shivashankara AR, et al. The health benefits of Indian traditional ayurvedic Rasayana (Anti-Aging) drugs: a review. Foods and Dietary Supplements in the Prevention and Treatment of Disease in Older Adults. 2015:151-61. CrossRef
- 5. Basudkar V, Gujrati G, Ajgaonkar S, et al. Emerging vistas for the nutraceutical withania somnifera in inflammaging. Pharmaceuticals (Basel). 2024;17(5):597. CrossRef PubMed
- 6. Vaidya VG, Naik NN, Ganu G, et al. Clinical pharmacokinetic evaluation of Withania somnifera (L.) Dunal root extract in healthy human volunteers: a non-randomized, single dose study utilizing UHPLC-MS/MS analysis. J Ethnopharmacol. 2024;322:117603. CrossRef PubMed
- 7. Verpoorte DR. New times for traditional medicine research. J Ethnopharmacol. 2017;197(197):1. CrossRef PubMed
- 8. Goyache I, Yavorov-Dayliev D, Milagro FI, et al. Caenorhabditis elegans as a screening model for probiotics with properties against metabolic syndrome. Int J Mol Sci. 2024;25(2):1321. CrossRef PubMed
- 9. Xu S, Hsiao TI, Chisholm AD. The wounded worm: using C. elegans to understand the molecular basis of skin wound healing. In: Worm 2012 (Vol. 1, No. 2, pp. 134-138). Taylor & Francis. CrossRef
- 10. Sifri CD, Begun J, Ausubel FM. The worm has turned – microbial virulence modeled in Caenorhabditis elegans. Trends Microbiol. 2005;13(3):119-127. CrossRef PubMed
- 11. Arya U, Das CK, Subramaniam JR. Caenorhabditis elegans for preclinical drug discovery. Curr Sci. 2010;1669-1680. Online
- 12. Kojima T, Kamei H, Aizu T, et al. Association analysis between longevity in the Japanese population and polymorphic variants of genes involved in insulin and insulin-like growth factor 1 signaling pathways. Exp Gerontol. 2004;39(11-12):1595-1598. CrossRef PubMed
- 13. Suh Y, Atzmon G, Cho MO, et al. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc Natl Acad Sci USA. 2008;105(9):3438-3442. CrossRef PubMed
- 14. Kumar R, Gupta K, Saharia K, et al. Withania somnifera root extract extends lifespan of Caenorhabditis elegans. Ann Neurosci. 2013;20(1):13-16. CrossRef PubMed
- 15. United States Pharmacopeial Convention. Ashwagandha root; powdered ashwagandha root; and powdered ashwagandha root extract, U.S. Pharmacopeia, N. Formulary (Eds.), Dietary supplements compendium. Rockville, MD, 2019.
- 16. Corsi AK, Wightman B, Chalfie M. A transparent window into biology: a primer on Caenorhabditis elegans. Genetics. 2015;200(2):387-407. CrossRef PubMed
- 17. Hamid R, Rotshteyn Y, Rabadi L, et al. Comparison of alamar blue and MTT assays for high through-put screening. Toxicol In Vitro. 2004;18(5):703-710. CrossRef PubMed
- 18. Tritten L, Braissant O, Keiser J. Comparison of novel and existing tools for studying drug sensitivity against the hookworm Ancylostoma ceylanicum in vitro. Parasitology. 2012;139(3):348-357. CrossRef PubMed
- 19. Bhalani DV, Nutan B, Kumar A, et al. Bioavailability enhancement techniques for poorly aqueous soluble drugs and therapeutics. Biomedicines. 2022;10(9):2055. CrossRef PubMed
- 20. Mitchell DH, Stiles JW, Santelli J, et al. Synchronous growth and aging of Caenorhabditis elegans in the presence of fluorodeoxyuridine. J Gerontol. 1979;34(1):28-36. CrossRef PubMed
- 21. Aitlhadj L, Stürzenbaum SR. The use of FUdR can cause prolonged longevity in mutant nematodes. Mech Ageing Dev. 2010;131(5):364-365. CrossRef PubMed
- 22. Hardaker LA, Singer E, Kerr R, et al. Serotonin modulates locomotory behavior and coordinates egg-laying and movement in Caenorhabditis elegans. J Neurobiol. 2001;49(4):303-313. CrossRef PubMed
- 23. Nagy S, Huang YC, Alkema MJ, et al. Caenorhabditis elegans exhibit a coupling between the defecation motor program and directed locomotion. Sci Rep. 2015;5(1):17174. CrossRef PubMed
- 24. Zavagno G, Raimundo A, Kirby A, et al. Rapid measurement of ageing by automated monitoring of movement of C. elegans populations. Geroscience. 2024;46(2):2281-2293. CrossRef PubMed
- 25. Longhin EM, El Yamani N, Rundén-Pran E, et al. The alamar blue assay in the context of safety testing of nanomaterials. Front Toxicol. 2022;4:981701. CrossRef PubMed
- 26. Banse SA, Lucanic M, Sedore CA, et al. Automated lifespan determination across Caenorhabditis strains and species reveals assay-specific effects of chemical interventions. Geroscience. 2019;41:945-960. CrossRef
- 27. Bansal A, Zhu LJ, Yen K, et al. Uncoupling lifespan and healthspan in Caenorhabditis elegans longevity mutants. Proc Natl Acad Sci USA. 2015;112(3):E277-E286. CrossRef PubMed
- 28. Ravi B, Garcia J, Collins K. The HSN egg-laying command neurons regulate the defecation motor program in Caenorhabditis elegans: integration. MicroPubl Biol. 2019;2019. PubMed
- 29. Klass MR. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech Ageing Dev. 1977;6(6):413-429. CrossRef PubMed
- 30. Pickett CL, Dietrich N, Chen J, et al. Mated progeny production is a biomarker of aging in Caenorhabditis elegans. G3 (Bethesda). 2013;3(12):2219-2232. CrossRef PubMed
- 31. Herndon LA, Schmeissner PJ, Dudaronek JM, et al. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419(6909):808-814. CrossRef PubMed
- 32. Ruegenberg S, Horn M, Pichlo C, et al. Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis. Nat Commun. 2020;11(1):687. CrossRef PubMed
- 33. Denzel MS, Storm NJ, Gutschmidt A, et al. Hexosamine pathway metabolites enhance protein quality control and prolong life. Cell. 2014;156(6):1167-1178. CrossRef PubMed