 |
Drug Target Insights 2025; 19: 1-10 ISSN 1177-3928 | DOI: 10.33393/dti.2025.3171 ORIGINAL RESEARCH ARTICLE |
 |
Characterization and enhanced antibiofilm activity of Annona muricata extract in combination with fluconazole against Candida albicans
ABSTRACT
Introduction: Candida albicans biofilm formation is a significant contributor to antifungal resistance, necessitating new treatment strategies. Annona muricata Lin., a traditional herbal remedy, has shown promise in combating microbial infections. The purpose of this study was to assess the antibiofilm activity of the methanol extract of A. muricata leaves alone or with the addition of fluconazole against C. albicans.
Methods: Phytochemicals from the methanol extract were analyzed by LC-MS, the XTT assay was used for metabolic activity, and morphological characteristics were examined using scanning electron microscopy (SEM). Molecular docking screening of identified compounds in A. muricata methanol leaves extract against a Sap3 receptor (PDB: 2H6T) was also performed.
Results: The LC-MS analysis detected 17 possible phytochemicals. The methanol extract showed a dose-dependent inhibition of biofilm formation, with maximum inhibition of ~60% observed at 240 μg/ml, and inhibition by fluconazole increased from 32% to 76% as the concentration increased from 15 to 240 μg/ml. The combination of A. muricata and fluconazole increased the inhibition significantly, from 74% to 78% at 15 μg/ml to 240 μg/mL, respectively. SEM of control and treated C. albicans biofilms showed an altered morphology and loss of cell integrity by the combination, corroborating the findings. Plant phytochemicals also possess high binding affinity (−9.7 to 8.0 kcal/mol, respectively) for the Sap3 enzyme and may therefore have therapeutic potential against C. albicans.
Conclusion: Consequently, the findings indicate that compounds in the A. muricata methanol extract may function in concert with fluconazole at sub-inhibitory concentrations to suppress C. albicans biofilm formation. This finding paves the way for the formulation and development of antifungal treatment regimens that may limit the development of fluconazole resistance employing this plant part.
Keywords: A. muricata, Candida albicans, Fluconazole, LC-MS, Synergism, XTT assay
Received: June 17, 2024
Accepted: December 4, 2024
Published online: January 13, 2025
This article includes supplementary material
Corresponding author:
Abhay P. Mishra
email: abhaypharmachemhnbgu@gmail.com
Drug Target Insights - ISSN 1177-3928 - www.aboutscience.eu/dti
© 2024 The Authors. This article is published by AboutScience and licensed under Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0).
Commercial use is not permitted and is subject to Publisher’s permissions. Full information is available at www.aboutscience.eu
Introduction
Seventy-five percent of human microbial infections are caused by the growth and persistence of biofilms, which are surface-attached microbial populations encased in a self-synthesized polymeric matrix (1). The ability of Candida albicans to form biofilms is a crucial component of its pathogenicity since it can increase tolerance to the host immune system and traditional antifungal medication (2). Most severe and recurring infections caused by C. albicans are linked to the development of biofilms on natural or synthetic surfaces (3). C. albicans biofilm has been known to cause persistent infections of organs and tissues invasively via seeding disseminated bloodstream infections, known as candidemia (4,5). Interestingly, secreted aspartyl proteinases (Saps) are among the hydrolytic enzymes that contribute significantly to the pathogenicity of the opportunistic pathogen C. albicans (6). It is believed that mucosal infections are associated with Sap 1-3, while systemic infections are associated with Sap 4-6 (7).
Herbal remedies have been utilized for millennia to cure a wide range of illnesses, both in Africa and elsewhere. This is mostly due to the perception that medicinal herbs are more accessible, more affordable, and more effective than Western medications (8). Annona muricata Lin. (Custard-apple, Annonaceae) is endemic to the West Indies and Central America, where it grows widely at elevations below 900 meters above sea level. It is also known by the names guanabana, paw-paw, soursop, and graviola. It grows in nations such as India, Angola, Puerto Rico, Brazil, Costa Rica, Colombia, and Venezuela that have tropical or subtropical climates (9). It has proven possible to isolate >200 bioactive chemicals from the A. muricata. Terpenoids, phenolics, and alkaloids are the most identified secondary metabolites of A. muricata (9-11). In Africa, India, and tropical America, A. muricata is frequently used as a folk remedy for a variety of human illnesses, including diabetes, rheumatism, cancer, and parasitic infections (12). The antibacterial, antifungal, anticancer, anticonvulsant, sedative, antiparasitic, and cardio-depressant properties of A. muricata leaves are just a few of its many potential uses (13,14). According to Rustanti and Fatmawati, A. muricata leaves ethanol extract showed antifungal activity, particularly against C. albicans (13). However, the literature presently has inadequate proof of the use of A. muricata leaf extract on biofilms of C. albicans.
Thus, this work aims to examine the antifungal activity and synergistic potential of the methanol extract of A. muricata leaf when combined with an antifungal agent, fluconazole, against C. albicans biofilm. Also, the existence of putative chemical components of A. muricata leaf extract, as well as the possibility of an interaction between identified compounds of A. muricata leaf and the Sap3 C. albicans, will be investigated to find new inhibitor candidates.
Experimental
Plant material collection
The A. muricata leaves were gathered in August 2020 at Mabira Forest in Uganda’s Buikwe District, with Voucher specimen No. AMHa4567, the plant was authenticated and confirmed by a taxonomist at the Makerere University Herbarium, Department of Plant Science, Microbiology and Biotechnology, Makerere University, Uganda.
Extraction procedure
Briefly, properly washed, cleaned, and dried (21 days at room temperature), leaves were ground into fine powder by using an electrical grinder. Powdered leaves (1000 g) were extracted successively (2.5 L × 3) by macerating with n-hexane, dichloromethane, ethyl acetate, methanol, and water at room temperature for 72 hours to obtain four respective extracts. The extracts were filtered through cotton wool and then dried in a rotary evaporator set at 40°C. To remove any remaining moisture, the extracts were moved to sample bottles and put in a desiccator with anhydrous sodium sulfate. The dried extracts were then refrigerated at −20°C in t-stopped bottles for further analysis (15). According to the studies, polar solvents are better than non-polar solvents, most likely because they offer a higher phytocompound recovery yield (16,17). Therefore, the adoption of methanol extract for additional research was motivated by the increased extraction yield.
LC-MS parameters for secondary phytochemical analysis
An ABSCIEX 4000 QTRAP hybrid triple quadrupole ion trap mass spectrometer was used to analyze the samples, and its front end was a Shimadzu HPLC stack. The software Analyst 1.5 (AB SCIEX) was used for all data processing and collecting. A 20 μL sample was separated using a 10-minute gradient that started with 5% solvent A (H2O containing 0.1% ammonium formate) and gradually progressed to 10% solvent B (MeOH with 0.1% ammonium formate). This separation was conducted on a C18 column (150 × 4.6 mm, Discovery, Supelco) at a flow rate of 0.5 mL/min. The solvent gradient was gradually increased to 30% B over the next 10 minutes, then to 50% B for an additional 10 minutes, and finally to 95% B over the subsequent 15 minutes. The total runtime for the experiment was 60 minutes, which allowed for column re-equilibration. Analytes that were eluted were electrosprayed into the TurboV ion source at 500°C to evaporate any excess solvent. The system used a nebulizer gas pressure of 30 psi, a heater gas pressure of 30 psi, and a curtain gas pressure of 20 psi. The ion spray voltage was adjusted to 5500V in positive ionization mode and −4500V in negative ionization mode. With fixed declustering and entrance potentials of 40 and 10V, respectively, the eluting analytes were mass measured on the mass spectrometer in Q1 scan mode, covering a range of 100 Da to 2000 Da for a 3-second cycle period.
Multiple optimizations and scans were done to reduce the background noise and come up with the best baseline stabilizations for accurate compound identifications.
The compounds were identified by comparing the outcomes of ESI-MS/MS observations articulated as tR and fragmentation configurations with those disclosed by investigations obtained from various database systems, such as the National Library of Medicine, Mass Bank of Europe, and related literature reviews. MS spectra, TIC, and possible identified compounds with their corresponding structures were retrieved from the library as well as the PubChem Compounds database.
Docking Studies
Using the Lamarckian genetic algorithm (LGA) as the scoring function, the PyRx virtual screening program was utilized to virtually screen ligands, including AutoDock and AutoDock Vina. Molecular docking analysis was used to determine the chemical compounds’ binding affinity with the examined 2H6T protein’s active site amino acids. The SDF format of Sap3 (PDB ID: 2H6T) protein 3D structure was downloaded from the protein data bank (PDB). The AutoDock tool was used to import the proteins with access codes 2H6T. After the water molecules attached to the structures were eliminated, hydrogen atoms were inserted. Similarly, LCMS-identified compound structures were also downloaded from the PubChem database. The ligands were docked using the active site dimensions, which were defined as a grid size based on the XYZ axis. A maximum exhaustiveness of 10 was computed for every ligand. Prior to the docking process starting, AutoDock Vina applied charges to the structures of proteins and ligands (15,18). The 2D visualization of the ideal binding pose—which is the binding energy with the lowest value for each compound-protein combination—was studied using BIOVIA Discovery Studio software. Hence, there is a great chance that the ligands produced will be employed as medication candidates.
Antibiofilm activity
Strains used
For this investigation, C. albicans SC5314 was revived and maintained on Yeast Malt extract (YM) (10 g/L glucose, 3 g/L yeast extract, 3 g/L malt extract, 5 g/L peptone, and 16 g/L agar) agar plates and preserved at 4°C for all studies conducted.
XTT assay of biofilms
On YM agar plates, C. albicans cells were cultured and subsequently incubated for 24 hours at 30°C. Following incubation, 20 mL of yeast nitrogen base (YNB) glucose medium (10 g/L glucose, 6.7 g/L YNB) was filled with a loop full of cells, and the mixture was incubated for 24 hours at 30°C.
Using an Eppendorf centrifuge 5430R, cells were harvested and washed (3075 g × 3 for 5 minutes at 4°C) with phosphate-buffered saline (PBS) and resuspended in 10 mL of RPMI-1640 media (Sigma-Aldrich, UK) at a concentration of 1x10–6 cells/ml. Aliquots of 200 µL of the cell suspension, including A. muricata methanol extract (reconstituted in sterile water) (AM), fluconazole (FLU) and a combination of the extract and antifungal drug (AM + FLU) at final concentrations ranging from 15-240 µg/ml was dispensed into a 96-well microtiter plate (Corning Incorporated, Costar®, U.S.) and incubated for 48 hours at 37°C to allow biofilm formation. Standardized cells with RPMI-1640 media were used as the negative control. To get rid of non-adherent cells, wells were twice cleaned with 200 µL PBS after 48 hours. Kuhn et al.’s (19) method of assessing the mitochondrial metabolic activity of the biofilms was used for the reduction of 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)carbonyl]-2H-tetrazolium hydroxide (XTT) (Sigma Aldrich, UK) in order to assess the viability of the yeast.
In the cellular supernatant, XTT was converted to colored, diffusible, water-soluble formazan, whose optical density was easily measured at 492 nm. Three biological duplicates of each three technical duplicates were used for this experiment. %Inhibition was calculated according to the following formula:
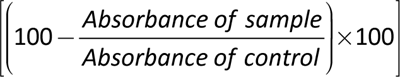
After calculating the average and standard deviations, the student’s t-test was performed to assess the significance of the data sets, with P < 0.05 being deemed significant. IC50 values were determined by performing non-linear fitting analysis using GraphPad Prism 8.0.2 software.
Scanning electron microscopy (SEM)
Biofilm was prepared as described above in a flat bottom 6-well plate (Greiner Bio-One, Germany) in 2 mL of RPMI-1640 medium containing either FLU (15 µg/mL), AM (15 µg/mL) or a combination of FLU (7.5 µg/mL) and AM (7.5 µg/mL) on sterile polymer discs (Isopore 0.2 µm hydrophilic polycarbonate membrane disc, Merck, Germany). The polymer discs were removed aseptically after incubation and left overnight in a primary fixative solution of 3% (v/v) glutardialdehyde (Merck, Germany) in phosphate buffer (pH 7.0). The biofilms underwent two PBS washes before being fixed for two hours at room temperature using 1% (v/v) osmium tetroxide (Merck, Germany) as a secondary fixative. This was followed by a second wash phase. The biofilms were air dried in a desiccator after being dehydrated in an ethanol series (50% for 20 minutes, 70% for 20 minutes, 95% for 20 minutes, and 100% for one hour – twice). Then, they were subsequently coated with gold using SEM coating equipment (EM ACE600, Leica, Austria) and exposed to critical point drying (Samdri®-795 Critical Point Dryer, Tousimis, United States of America) for 30 minutes. Biofilms were analyzed using a JSM-7800F field emission SEM (ZEISS, Germany).
Results and Discussion
LCMS profile of A. muricata methanol extract
A total of 17 phytochemicals were detected by LC/MS and 14 among them were identified (Table 1). These included alkaloids (asimilobine 2, swainsonine 3, xylopine 7, (+)-4’-O-methylcoclaurine 10), flavonoids (isovitexin 4, vitexin 5, rutin 8, quercetin-3-O-D-glucuronide 9, kaempferol 3-O-rutinoside 11, eriocitrin 15, apigenin 6,8-di-C-glucoside 17) and acetogenins (annonisin 13, annomuricin A 14, montanacin B/C 16) (Fig. 1) from AM (Fig. 1). A potential identification of the chemicals was predicated on contrasting the fragmentation patterns and retention times (tR) obtained from ESI-MS/MS experiments with those documented in research gathered from various databases.
The first-order mass spectrum of compound 3 showed an intense protonated molecular ion [M+H]+ at m/z 174.3 and also showed a single loss of one hydroxyl group as water [M+H-H2O]+ at m/z at 156.2, which was identified as swainsonine (20-22). A similar trend was observed for compound 2, where a protonated molecular ion [M+H]+ was observed at m/z 268.6. The fragments at m/z 250.6 and 236.5 were assigned to the loss of the hydroxyl group as water [M+H−H2O]+ and methoxy group [M+H−OCH3]+, respectively, and this led to the identification of compound 2 as asimilobine and this was consistent with the results of Lima et al. and Mohanty et al. (23,24). Compound 7 was identified as xylopine based on the typical fragment ions at m/z 279.4 ([M + H−OH]+) and 249.7 ([M−CH2O2]+) (22,25,26). For compound 10, the main fragment ion appeared at m/z 300.4 [M+H]+ and another main fragment at m/z 283.9 [M−OH]+, which corresponded to the loss of the hydroxyl group. Moreover, other fragments at m/z 251.8, 178.3, and 121.1 were observed, which led to the identification of compound 10 as (+)-4’-O-Methylcoclaurine (27).
| Peak | Compound name | tR (min) | m/z | Ionization mode | Molecular formula | References |
|---|---|---|---|---|---|---|
| 1 | Unidentified | 3.585 | 161.0 | [M-H]– | – | – |
| 2 | Asimilobine | 4.749 | 268.6 | [M+H]+ | C8H15NO3 | (24) |
| 3 | Swainsonine | 9.286 | 174.3 | [M+H]+ | C17H17NO2 | (20-22) |
| 4 | Isovitexin | 12.663 | 431.0 | [M-H]– | C21H20O10 | (32) |
| 5 | Vitexin | 13.832 | 431.0 | [M-H]– | C21H20O10 | (32) |
| 6 | Unidentified | 16.112 | 407.0 | [M-H]– | – | – |
| 7 | Xylopine | 18.298 | 296.2 | [M+H]+ | C20H21NO4 | (22,25,26) |
| 8 | Rutin | 17.381 | 609.7 | [M-H]– | C27H30O16 | (30) |
| 9 | Quercetin-3-O-D-glucuronide | 17.429 | 477.1 | [M+H]– | C21H20O13 | (33) |
| 10 | (+)-4’-O-Methylcoclaurine | 16.889 | 300.4 | [M-H]+ | C18H21NO3 | (27) |
| 11 | Kaempferol 3-O-rutinoside | 19.900 | 595.3 | [M+H]+ | C27H30O15 | (28,29) |
| 12 | Unidentified | 22.840 | 517.4 | [M+H]– | – | – |
| 13 | Annonisin | 30.812 | 611.3 | [M+H]+ | C35H62O8 | (36) |
| 14 | Annomuricin A | 31.356 | 613.3 | [M+H]+ | C35H64O8 | (38,39) |
| 15 | Eriocitrin | 33.166 | 595.7 | [M+H]– | C27H32O15 | (31,34) |
| 16 | Montanacin B | 38.016 | 611.2 | [M+H]+ | C35H62O8 | (37) |
| 17 | Apigenin 6,8-di-C-glucoside | 38.177 | 593.4 | [M+H]– | C27H30O15 | (31) |
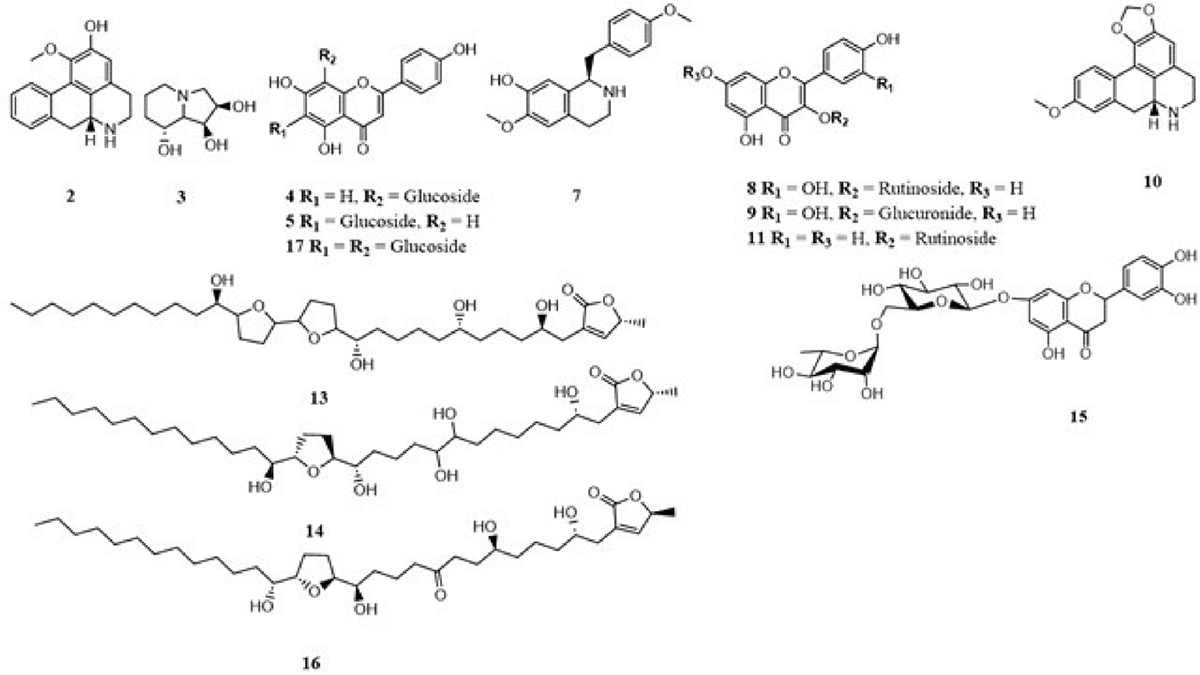
FIGURE 1 - Chemical structures of compounds identified from AM by LC-MS
The mass spectrum fragmentation of compound 11 appeared at m/z 595.3 [M+H]+. The product ion spectrum of the compound 11 ion (m/z 595.3) showed fragment ion at m/z 577.4 [M+H−H2O]+ due to loss of water molecule. The fragments at m/z 449.4 and 287.4 indicated the loss of rhamnose and glucose sugar units, and therefore, compound 11 was identified as kaempferol 3-O-rutinoside (28,29). Similarly, a deprotonated molecular ion [M-H]– was observed at m/z 609.7 for compound 8. The spectra showed the presence of the main fragments at m/z 463.2 [M-H-146]– and 301.1 [M - H-146-162]–, indicating the loss of two sugar units consisting of rhamnose and a pyranose, and this led to the identification of compound 8 as rutin (30). Compound 17 showed a pseudo molecular ion peak at m/z 593.4 [M−H]-. MS showed fragment ions at m/z 431.1 [M−H-162] and 269.0 [M−H-162], corresponding to the presence of the hexose nature of C-glycoside flavone. Moreover, from the observed base peak fragment at m/z 269.0, it could be concluded that compound 17 was apigenin 6,8-di-C-glucoside (31). Similarly, compounds 4 and 5 were assigned as isovitexin and vitexin, respectively (32).
Compound 9 gave a pseudo-molecular ion at m/z 477.1 [M-H]–. The MS/MS spectrum featured characteristic ion of m/z 301.1 [M-H-178]–, derived from the loss of glucuronide, and this led to the identification of compound 9 as quercetin 3-O-glucuronide (33). For compound 15, the LC-MS chromatogram yielded an intense peak at m/z 595.7 [M-H]–. Moreover, another main fragments at m/z 449.2 [M-H-146]– and 287.0 [M-H-162]– were observed, which implies the loss of deoxyhexose and hexose as rhamnose and a pyranose, and therefore, compound 15 was identified as eriocitrin (31,34).
For compounds 13 and 16, the LC-MS/MS chromatogram showed a molecular ion peak at m/z 611.2 [M+H]+, which corresponds to the molecular formula C35H62O8. Due to consecutive losses of water (18 Da), the mass fragments at m/z 575.4 [M+H-H2O]+, 557.4 [M+H-2H2O]+, 539.5 [M+H-3H2O]+, and 521.7 [M+H-4H2O]+ show the existence of four hydroxyl groups. This was in agreement with Gu et al. (35), who reported that fragment ions, indicative of the multihydroxylated structures of the acetogenins, were generated from consecutive losses of H2O (three to five molecules). This, therefore, led to the identification of compounds 13 and 16 as annonisin and montanacin B, respectively (36,37). Compound 14 gave a molecular ion peak at m/z 613.35 [M+H]+, which corresponds to the molecular formula C35H64O8 (38). Similarly to compound 13 and 16, compound 14 also had the same fragments at 577.4 [M+H-H2O]+, 559.2 [M+H-2H2O]+, 541.5 [M+H-3H2O]+, 523.6 [M+H-4H2O]+ and 505.6 [M+H-5H2O]+ due to successive losses of water (18 Da) which indicates the presence of five hydroxyl groups. These results were in agreement with the literature (38,39), and compound 14 was identified as annomuricin A.
Molecular docking
In order to produce docking scores that show the bio-affinity of the docked molecules, the molecular docking algorithm simulates ligand interactions in the target proteins’ or receptors’ active regions (40). C. albicans is reported to have Sap 1-10 genes. The Sap proteins are one of the classic pathogenic factors whose expression is controlled by numerous parameters such as pH levels, temperature, location of infection, and physicochemical ambient conditions. Since Sap3 is implicated in mucosal infections, the development of inhibitors targeting Sap3 is a promising strategy for addressing infections caused by C. albicans (7).
The molecular docking calculations presented in Table 2 indicate that the enzyme Sap3 (2H6T) exhibited the utmost binding affinities with eriocitrin (-9.7 kcal/mol). The 2D visualization (Fig. 2A) displayed that eriocitrin established strong hydrogen bonds with GLY 34, ASP 86, SER 36, and ASN 192 at the protein active site. However, it also used a distinct pi bond to engage with ASP 218 at the catalytic pocket of the enzyme. In addition, the 2H6T macromolecule interaction with apigenin 6,8-di-C-glucoside showed the second-highest binding energy (−8.9 kcal/mol) through strong hydrogen bonds with GLY 220, THR 222, THR 300, and TYR 225 along with ASP 86 (π bond), GLY 85 (π-π stacked), as well as a noncovalent (amide-π stacking) interaction with TYR 225 (Fig. 2B). The strength of binding between receptors and ligands is determined by the number of hydrogen bonds produced. Also, binding energy influences the efficiency of ligand binding to enzymes (7).
| S. No. | Compound Name | Macromolecule | Binding energy (kcal/mol) | Closest residues of docked ligands at the active site of macromolecule |
|---|---|---|---|---|
| A. | Eriocitrin | 2H6T | –9.7 | GLY 34, ASP 86, SER 36, ASN 192, ASP 218 |
| B. | Apigenin 6,8-di-C-glucoside | 2H6T | –8.9 | GLY 220, THR 222, TYR 225, ASP 86, GLY 85, TYR 225 |
| C. | Xylopine | 2H6T | –8.5 | GLY 220, GLY 85, TYR 225, ASP 218, ILE 305 |
| D. | Isovitexin | 2H6T | –8.5 | ASP 86, THR 222, VAL 12, ASP 218, ILE 305 |
| E. | Rutin | 2H6T | –8.4 | ASP 218, GLY 85, GLY 34, THR 221, ILE 82 |
| F. | Vitexin | 2H6T | –8.1 | ASN 35, ILE 82, GLU 83, GLY 34, |
| G. | Asimilobine | 2H6T | –8.1 | ASP 32, 86, 218, THR 221, 222, ILE 123, 305, TYR 84, 225, GLY 34, 85, 220, SER 35 |
| H. | Kaempferol 3-O-rutinoside | 2H6T | –8.0 | TYR 84, SER 35, GLU 83, GLY 34, 85, ILE 82, TYR 303 |
| I. | (+)-4’-O-Methylcoclaurine | 2H6T | –7.8 | ILE 123, 305, TYR 84, 303, GLY 34, 85, 220, ASP 32, 86, 218, VAL 30, THR 221, SER 13 |
| J. | Quercetin-3-O-D-glucuronide | 2H6T | –7.3 | ILE 123, TYR 221, GLY 85, 220, ASP 218 |
| K. | Annonisin | 2H6T | –7.2 | ILE 82, 123, SER 13, 35, VAL 12, 30, ASP 32, 218, THR 221, GLY 34, 85, 220, TYR 84, 303, GLU 83 |
| L. | Annomuricin A | 2H6T | –6.8 | ILE 123, 305, SER 13, VAL 12, 30, ASP 32, 86, 218, THR 221, 222, GLY 34, 85, 220, TYR 84, 225, 303, GLU 83 |
| M. | Montanacin B | 2H6T | –6.7 | ILE 123, 305, SER 13, VAL 12, 30, ASP 32, 86, 218, THR 221, 222, GLY 34, 85, 220, TYR 84, 225, 303 |
| N. | Montanacin C | 2H6T | –6.4 | ILE 82, 123, 305, VAL 12, 30, ASP 32, 86, 218, GLY 34, 85, 220, TYR 84, 303, GLU 83 |
| O. | Swainsonine | 2H6T | –5.6 | GLY 34, SER 35 |
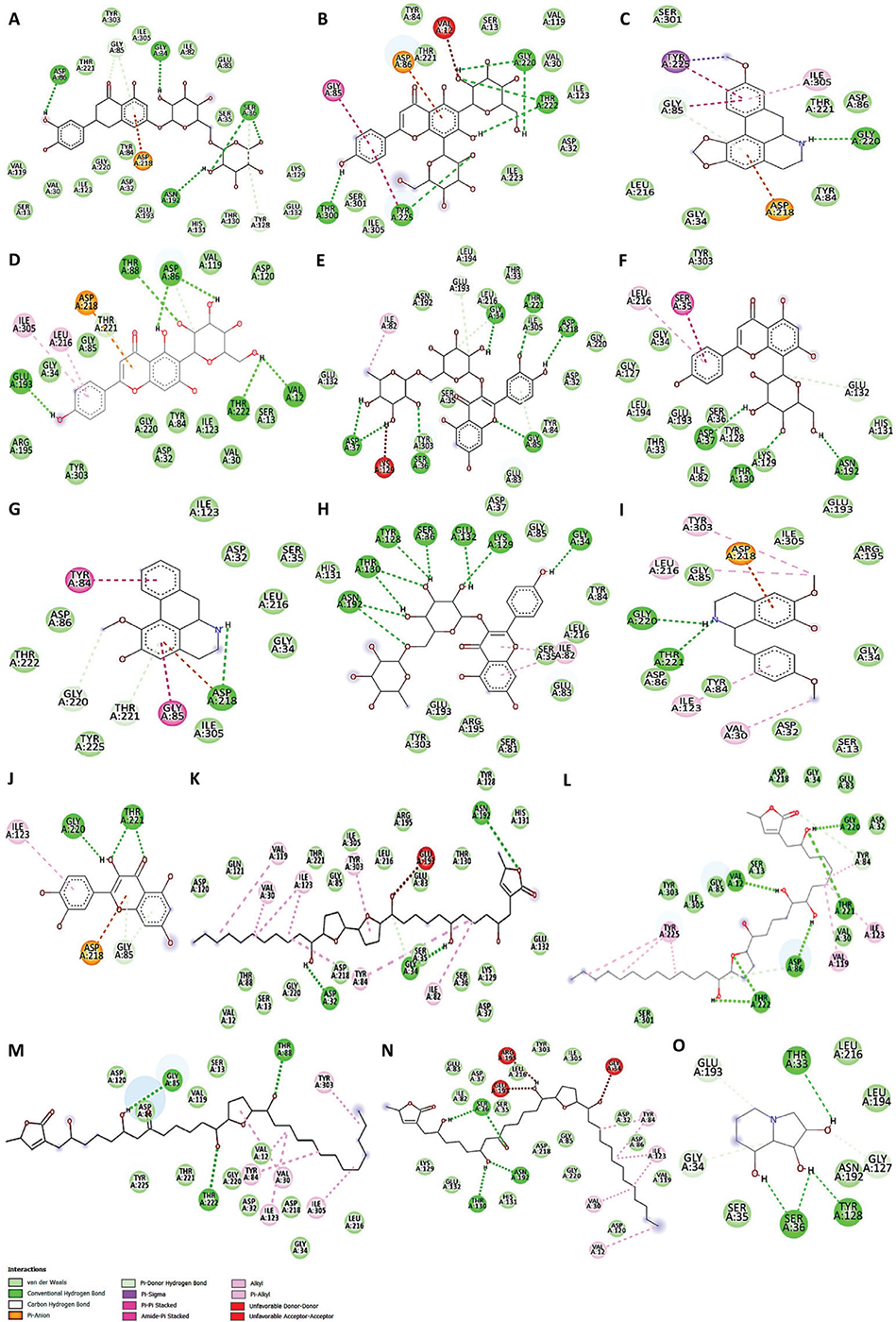
FIGURE 2 - 2D views of Sap3 active site amino acid residues interactions with A. muricata identified phytochemicals
Another molecule, xylopine, also formed bonds with GLY 220 (H-bond), GLY 85 (π stacking), and TYR 225 (π-σ) with a binding energy of −8.5 kcal/mol (Fig. 2C). Whereas isovitexin-2H6T showed same binding energy as xylopine, by interacting with GLU 193, THR 88, ASP 86, THR 222 and VAL 12 amino acids through hydrogen bonds and with LEU 216 by π-alkyl interactions (Fig. 2D). Intriguingly, the 2D viewpoint in Fig. 2C-D confirmed that the ASP 218 and ILE 305 amino acids are the brush border enzymes involved in π-anion and π-alkyl bond formation with both compounds, respectively. A noteworthy interaction has also been seen between rutin-2H6T molecular docking (binding energy of –8.4 kcal/mol). In such interactions, conventional hydrogen bonding consisted of ASP 37, ASP 218, SER 36, GLY 85, GLY 34, and THR 221, while alkyl bond formation occurred with ILE 82 amino acid (Fig. 2E). Vitexin interacted with 2H6T at active sites via ASN 192, SER 36, 35, 81, ILE 82, GLU 83, 132, 193, LEU 194, 216, GLY 34, HIS 131, THR 130, ASP 37, and LYS 129 amino acids with binding energy of −8.1 kcal/mol (Fig. 2F) whereas asimilobine interacted with same binding energy thru ASP 32, 86, 218, THR 221, 222, ILE 123, 305, TYR 84, 225, GLY 34, 85, 220, SER 35, LEU 216 amino acids interaction (Fig. 2G). Kaempferol 3-O-rutinoside interacted with 2H6T protein active sites through ASN192, THR 130, TYR 84, 128, SER 35, 36, 81, GLU 83, 132, 193 LYS 129, GLY 34, 85, ASP 37, HIS 131, LEU 216, ILE 82, ARG 195, and TYR 303 amino acids with binding energy of –8.0 kcal/mol (Fig. 2H). Other possible identified compounds such as (+)-4’-O-methylcoclaurine, quercetin-3-O-D-glucuronide, annonisin, annomuricin A, montanacin B, and montanacin C showed binding energy < –8.0 kcal/mol (Table 2) by interacting with several amino acids as shown in Fig. 2I-N.
In addition, with a binding value of –5.6 kcal/mol, swainsonine likewise showed strong attraction for the 2H6T catalytic site by interacting with GLU 193, THR 33, LEU 216, 194, GLY 34, 127, ASN 192, TYR 128, SER 35, 36 amino acid residues (Fig. 2O). All the ligands are docked inside the active site. As previously documented, the phytochemicals found in A. muricata exhibit potential antifungal activity against C. albicans (12,38,41,42). Thus, it is possible to infer that the inhibition of the Sap3 enzyme by the extract of A. muricata leaves (Fig. 3) is caused by the phytochemicals in the plant influencing the activity of the enzymes.
Effects of AM and AM+FLU on C. albicans biofilm formation
To determine if the predicted interaction may indeed cause an effect on biofilm formation of C. albicans, an in vitro antibiofilm assay was performed. The methanolic extract of A. muricata leaves showed a dose-dependent effect over C. albicans biofilm formation, as shown in Fig. 3.
The maximum level of inhibition observed was ~60% at 240 mg/mL. The biofilm inhibitory activities of a clinical antifungal drug used for the treatment of invasive fungal infections, fluconazole, were also evaluated in this study as positive control and showed a similar dose-dependent response, with ~70% inhibition observed at 120 mg/mL. Interestingly, at lower concentrations, the combined effect of the extract and fluconazole achieved synergistically increased levels of inhibition. At 15 mg/mL of the combination, biofilm formation was inhibited in excess of 70%. Our findings support Campos et al.’s 2023 investigation, which found that an ethanolic extract from A. muricata leaves has antifungal properties against a multidrug-resistant strain of C. albicans (38,42).
This synergism is also supported by the IC50 values (Suppl. Table 1). As expected, FLU (IC50 = 18.33 μg/mL) had a lower IC50 value than the AM (IC50 = 128.70 μg/mL). Importantly, the IC50 value of the combination (IC50 = 0.83 μg/mL) is two orders of magnitude lower than fluconazole, demonstrating a pronounced synergistic activity. A low IC50 value indicates that the drug is effective at lower concentrations, resulting in reduced systemic toxicity when administered to patients (43). Identically, it has been seen that combined treatment is a proposed approach to overcome the issues associated with single-agent therapy; numerous studies have shown that combination therapies are superior to single-agent therapy (44,45). Accordingly, flavonoids (such as quercetin, catechin, epigallocatechin gallate, etc.) are a promising synergistic agent when combined with fluconazole and are an effective antifungal agent against C. albicans biofilms (45-47). Therefore, the phytocomponents of A. muricata leaves may play a role in their promising antifungal properties and possibly produce a synergistic effect with fluconazole.
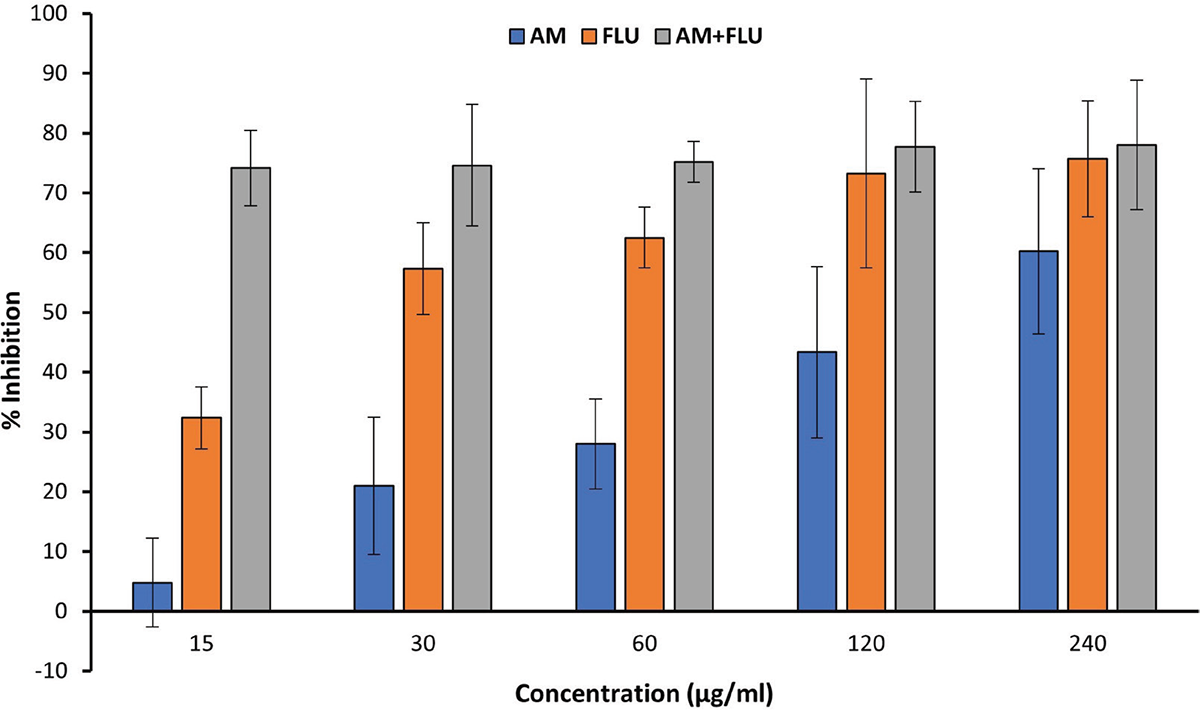
FIGURE 3 - Percent inhibition of C. albicans biofilms formation by A. muricata ethanol extract (AM), fluconazole (FLU), and the combination (AM + FLU, 1:1). All experiments were performed in biological and technical triplicates. The quantitative data are presented as mean ± standard deviation (SD).
Effect of AM and AM + FLU on the cell surface of C. albicans
Scanning electron micrographs of the control and treated (AM and AM+FLU) biofilms showed that untreated cells formed normal biofilms consisting of hyphae with smooth surfaces (Fig. 4A). As expected, the biofilms grown in the presence of FLU were less dense and hyphal formation was inhibited (Fig. 4B). Biofilms exposed to AM alone, were inhibited compared to the controls, but still displayed hyphal formation (Fig. 4C). In Fig. 4D, details of the biofilm exposed to the combination of fluconazole and extract can be seen. These biofilms were very sparse, and hyphal formation was inhibited. The release of cellular material can be seen. Studies have reported that A. muricata leaf extract causes modifications to the surface cells in biofilm cells by targeting the cell envelope of C. albicans. This effect may result from several identified compounds in the plant extract (38,42). We identified many of the same compounds reported previously (38), and there is a possibility that these compounds are responsible for generating a synergistic effect by interacting with fluconazole when AM + FLU is given as a combined treatment.
Conclusion
LC-MS analysis of methanol extract demonstrated that A. muricata contains several chemical constituents, 14 of which were identified. As indicated by molecular docking, some of these may interact with the secreted protease Sap3, which is an important hydrolytic enzyme of C. albicans. Our findings regarding the antibiofilm activity of AM leaf extract corroborate recent studies conducted by Campos and co-workers (38,42), who demonstrated that the ethanolic extract of A. muricata leaf is a promising anticandidal agent and can reduce fungal infection (in vitro and in vivo). In addition, we established that the combination of A. muricata methanol extract and fluconazole shows synergistic increased inhibition of biofilm formation, which may be advantageous in combating fluconazole resistance in C. albicans by lowering the effective dose required to inhibit biofilm formation. These results suggest that more research is necessary to fully understand the underlying mechanisms of action of AM + FLU and explore its potential as an anticandidal agent in vivo.
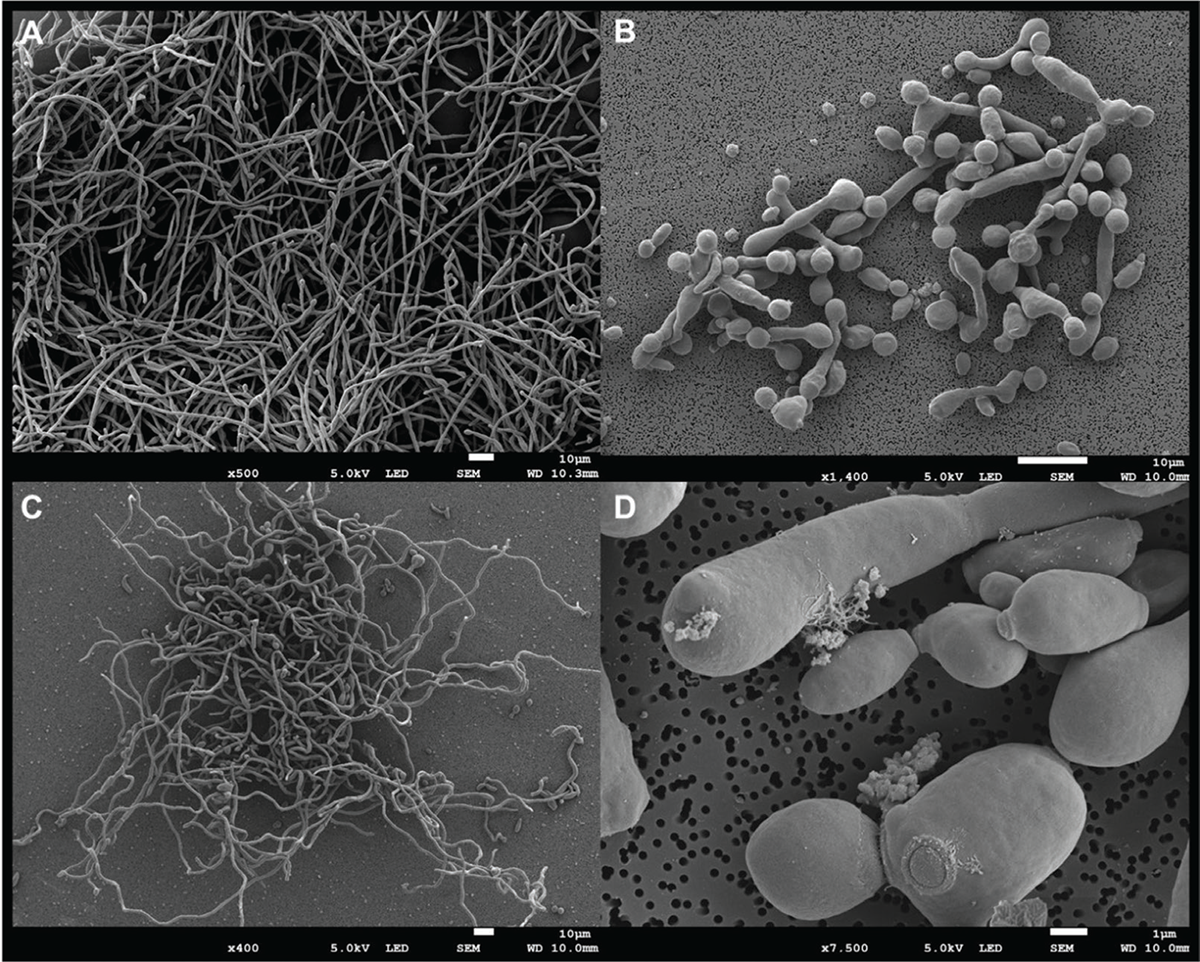
FIGURE 4 - Morphology of C. albicans biofilms grown on sterile polymer discs for 48 hours at 37°C. A. Control biofilms, B. Biofilms grown in the presence of 15 µg/mL FLU, C. Biofilms grown in the presence of 15 µg/mL AM, D. Detail of biofilm cells grown in the presence of both FLU (7.5 µg/mL) + AM (7.5 µg/mL).
Disclosures
Conflict of interest: The authors declare no conflict of interest.
Financial support: This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Data availability statement: The research data associated with this article are included within the article and in the supplementary material of this article.
Author’s contribution: Conceptualization: APM, CHP; Data Curation: APM, MY, JN; Formal Analysis: LKS, GK, APM, CHP, MY; Investigation: APM, LKS, MY, GK; Methodology: APM, LKS, MY, GK; Project Administration: APM, MGM, CHP; Supervision: CHP, MGM; Writing Original Draft: APM; Writing – Review & Editing: APM, GK, CHP.
References
- 1. Miquel S, Lagrafeuille R, Souweine B, et al; Anti-biofilm activity as a health issue. Front Microbiol. 2016;7:592. CrossRef PubMed
- 2. Wang S, Wang P, Liu J, et al; Antibiofilm activity of essential fatty acids against Candida albicans from Vulvovaginal candidiasis and bloodstream infections. Infect Drug Resist. 2022;15:4181-4193. CrossRef PubMed
- 3. Pereira R, Dos Santos Fontenelle RO, de Brito EHS, et al; Biofilm of Candida albicans: formation, regulation and resistance. J Appl Microbiol. 2021;131(1):11-22. CrossRef PubMed
- 4. Fox E, Nobile C. The Role of Candida albicans Biofilms in Human Disease. In: Dietrich L, Friedmann T, eds. Candida Albicans: Symptoms, Causes and Treatment Options. Nova Science Publishers; 2013:1-24. Online (Accessed June 2024)
- 5. Gulati M, Nobile CJ. Candida albicans biofilms: development, regulation, and molecular mechanisms. Microbes Infect. 2016;18(5):310-321. CrossRef PubMed
- 6. Kim JS, Lee KT, Bahn YS. Secreted aspartyl protease 3 regulated by the Ras/cAMP/PKA pathway promotes the virulence of Candida auris. Front Cell Infect Microbiol. 2023;13:1257897. CrossRef PubMed
- 7. Gholam GM, Firdausy IA, Artika IM, et al; Molecular docking: bioactive compounds of Mimosa pudica as an inhibitor of Candida albicans Sap 3. Current Biochemistry. 2023;10(1):24-37. CrossRef
- 8. Ng’uni TL, dos Santos Abrantes PM, McArthur C, Klaasen JA, Fielding BC. Evaluation of synergistic anticandidal activity of Galenia africana extract and fluconazole against Candida albicans and Candida glabrata. J Herb Med. 2022;32:100503. CrossRef
- 9. Delgado Y, Cassé C, Ferrer-Acosta Y, et al. Biomedical effects of the phytonutrients turmeric, garlic, cinnamon, graviola, and oregano: A comprehensive review. Appl Sci (Basel). 2021;11(18):8477. CrossRef
- 10. Agu KC, Okolie PN. Proximate composition, phytochemical analysis, and in vitro antioxidant potentials of extracts of Annona muricata (Soursop). Food Sci Nutr. 2017;5(5):1029-1036. CrossRef PubMed
- 11. Coria-Téllez AV, Montalvo-Gónzalez E, Yahia EM, et al; Annona muricata: A comprehensive review on its traditional medicinal uses, phytochemicals, pharmacological activities, mechanisms of action and toxicity. Arab J Chem. 2018;11(5):662-691. CrossRef
- 12. Arthi S, Ramani P, Rajeshkumar S. Green synthesis of Annona muricata mediated selenium nanoparticles and its antifungal activity against Candida albicans. J Popul Ther Clin Pharmacol. 2023;30(16). CrossRef
- 13. Rustanti E, Fatmawati Z. The active compound of soursop leaf extract (Annona muricata, L.) as anti-vaginal discharge (Fluor albus). IOP Conf Ser Earth Environ Sci. 2020;456(1):012071. CrossRef
- 14. Egbo CC, Igboaka DC, Uzor PF. Antimicrobial assay and GC-MS profile of the extract of the endophytic fungus from Annona muricata (Annonaceae) leaf. Trop J Nat Prod Res. 2024;8(3):6731-6735.
- 15. Mishra AP, Nigam M, Nambooze J, et al; GC-MS analysis, antioxidant, and antidiabetic properties of methanol extract of Annona muricata L. leaves – An in vitro and in silico study. Curr Org Chem. 2023;27(17):1531-1541. CrossRef
- 16. Chigayo K, Mojapelo PEL, Mnyakeni-Moleele S, et al; Phytochemical and antioxidant properties of different solvent extracts of Kirkia wilmsii tubers. Asian Pac J Trop Biomed. 2016;6(12):1037-1043. CrossRef
- 17. Ahmed AS, McGaw LJ, Elgorashi EE, et al; Polarity of extracts and fractions of four Combretum (Combretaceae) species used to treat infections and gastrointestinal disorders in southern African traditional medicine has a major effect on different relevant in vitro activities. J Ethnopharmacol. 2014;154(2):339-350. CrossRef PubMed
- 18. Adi PJ, Yellapu NK, Matcha B. Modeling, molecular docking, probing catalytic binding mode of acetyl-CoA malate synthase G in Brucella melitensis 16M. Biochem Biophys Rep. 2016;8:192-199. CrossRef PubMed
- 19. Kuhn DM, Balkis M, Chandra J, et al; Uses and limitations of the XTT assay in studies of Candida growth and metabolism. J Clin Microbiol. 2003;41(1):506-508. CrossRef PubMed
- 20. Gardner DR, Molyneux RJ, Ralphs MH. Analysis of swainsonine: extraction methods, detection, and measurement in populations of locoweeds (Oxytropis spp.). J Agric Food Chem. 2001;49(10):4573-4580. CrossRef PubMed
- 21. Egan MJ, Kite GC, Porter EA, et al; Electrospray and APCI analysis of polyhydroxyalkaloids using positive and negative collision induced dissociation experiments in a quadrupole ion trap. Presented at SAC 99, Dublin, Ireland, July 25–30, 1999. Analyst. 2000;125(8):1409-1414. CrossRef
- 22. Fofana S, Keita A, Balde S, et al; Alkaloids from leaves of Annona muricata. Chem Nat Compd. 2012;48(4):714. CrossRef
- 23. Lima BRd, Silva F, Soares ER, et al; Integrative approach based on leaf spray mass spectrometry, HPLC-DAD-MS/MS, and NMR for comprehensive characterization of isoquinoline-derived alkaloids in leaves of Onychopetalum amazonicum RE Fr. J Braz Chem Soc. 2020;31:79-89. CrossRef
- 24. Mohanty S, Hollinshead J, Jones L, et al; Annona muricata (Graviola): Toxic or therapeutic. Natural Product Communications. 2008;3(1):1934578X0800300107. CrossRef
- 25. Ferraz CR, Silva DB, Prado LCDS, et al; Antidiarrhoeic effect and dereplication of the aqueous extract of Annona crassiflora (Annonaceae). Nat Prod Res. 2019;33(4):563-567. CrossRef PubMed
- 26. Nugraha AS, Haritakun R, Lambert JM, et al; Alkaloids from the root of Indonesian Annona muricata L. Nat Prod Res. 2021;35(3):481-489. CrossRef PubMed
- 27. Matsushige A, Kotake Y, Matsunami K, et al; Annonamine, a new aporphine alkaloid from the leaves of Annona muricata. Chem Pharm Bull (Tokyo). 2012;60(2):257-259. CrossRef PubMed
- 28. Cárdenas C, Torres-Vargas JA, Cárdenas-Valdivia A, et al; Non-targeted metabolomics characterization of Annona muricata leaf extracts with anti-angiogenic activity. Biomed Pharmacother. 2021;144:112263. CrossRef PubMed
- 29. Nawwar M, Ayoub N, Hussein S, et al; A flavonol triglycoside and investigation of the antioxidant and cell stimulating activities of Annona muricata Linn. Arch Pharm Res. 2012;35(5):761-767. CrossRef PubMed
- 30. Moghadamtousi SZ, Fadaeinasab M, Nikzad S, et al; Annona muricata (Annonaceae): a review of its traditional uses, isolated acetogenins and biological activities. Int J Mol Sci. 2015;16(7):15625-15658. CrossRef PubMed
- 31. Du J, Zhong B, Subbiah V, et al; LC-ESI-QTOF-MS/MS profiling and antioxidant activity of phenolics from Custard Apple fruit and by-products. Separations. 2021;8(5):62. CrossRef
- 32. George VC, Kumar DR, Rajkumar V, et al; Quantitative assessment of the relative antineoplastic potential of the n-butanolic leaf extract of Annona muricata Linn. in normal and immortalized human cell lines. Asian Pac J Cancer Prev. 2012;13(2):699-704. CrossRef PubMed
- 33. Cioffi, E., Comune, L., Piccolella, S., Buono, M., & Pacifico, S. (2023). Quercetin 3-O-Glucuronide from Aglianico Vine Leaves: A Selective Sustainable Recovery and Accumulation Monitoring. Foods, 12(14), 2646. CrossRef PubMed
- 34. Li L, Feng X, Chen Y, et al; A comprehensive study of eriocitrin metabolism in vivo and in vitro based on an efficient UHPLC-Q-TOF-MS/MS strategy. RSC Adv. 2019;9(43):24963-24980. CrossRef PubMed
- 35. Gu ZM, Zhou D, Wu J, et al; Screening for Annonaceous acetogenins in bioactive plant extracts by liquid chromatography/mass spectrometry. J Nat Prod. 1997;60(3):242-248. CrossRef PubMed
- 36. Al Kazman BSM, Harnett JE, Hanrahan JR. Identification of Annonaceous acetogenins and alkaloids from the leaves, pulp, and seeds of Annona atemoya. Int J Mol Sci. 2023;24(3):2294. CrossRef PubMed
- 37. Hidalgo JR, Gilabert M, Cabedo N, et al; Montanacin-L and montanacin-K two previously non-described acetogenins from Annona montana twigs and leaves. Phytochem Lett. 2020;38:78-83. CrossRef
- 38. Campos LM, Lemos ASO, Diniz IOM, et al; Antifungal Annona muricata L. (soursop) extract targets the cell envelope of multi-drug resistant Candida albicans. J Ethnopharmacol. 2023;301:115856. CrossRef PubMed
- 39. Duret P, Hocquemiller R, Cavé A. Annonisin, a bis-tetrahydrofuran acetogenin from Annona atemoya seeds. Phytochemistry. 1997;45(7):1423-1426. CrossRef
- 40. Apeh VO, Adegboyega AE, Chukwuma IF, et al; An in silico study of bioactive compounds of Annona muricata in the design of ani-prostate cancer agent: MM/GBSA, pharmacophore modeling and ADMET parameters. Inform Med Unlocked. 2023;43:101377. CrossRef
- 41. Abdul-Hammed M, Olaide IA, Adegoke HM, et al; Antifungal activities of phytochemicals from Annona muricate (Sour Sop): molecular docking and chemoinformatics approach. Majalah Obat Tradisional. 2022;27(3):218. CrossRef
- 42. Campos LM, Silva TP, de Oliveira Lemos AS, et al; Antibiofilm potential of Annona muricata L. ethanolic extract against multi-drug resistant Candida albicans. J Ethnopharmacol. 2023;315:116682. CrossRef PubMed
- 43. Berrouet C, Dorilas N, Rejniak KA, et al; Comparison of drug inhibitory effects (IC50) in monolayer and spheroid cultures. Bull Math Biol. 2020;82(6):68. CrossRef PubMed
- 44. Duarte D, Vale N. Evaluation of synergism in drug combinations and reference models for future orientations in oncology. Curr Res Pharmacol Drug Discov. 2022;3:100110. CrossRef PubMed
- 45. Gao M, Wang H, Zhu L. Quercetin assists fluconazole to inhibit biofilm formations of fluconazole-resistant Candida albicans in in vitro and in vivo antifungal managements of vulvovaginal candidiasis. Cell Physiol Biochem. 2016;40(3-4):727-742. CrossRef PubMed
- 46. da Silva CR, de Andrade Neto JB, de Sousa Campos R, et al; Synergistic effect of the flavonoid catechin, quercetin, or epigallocatechin gallate with fluconazole induces apoptosis in Candida tropicalis resistant to fluconazole. Antimicrob Agents Chemother. 2014;58(3):1468-1478. CrossRef PubMed
- 47. Liu W, Li LP, Zhang JD, et al; Synergistic antifungal effect of glabridin and fluconazole. PLoS One. 2014;9(7):e103442. CrossRef PubMed