 |
Drug Target Insights 2024; 18: 54-69 ISSN 1177-3928 | DOI: 10.33393/dti.2024.3082 ORIGINAL RESEARCH ARTICLE |
 |
Deciphering the molecular mechanisms underlying anti-pathogenic potential of a polyherbal formulation Enteropan® against multidrug-resistant Pseudomonas aeruginosa
ABSTRACT
Objective: Anti-pathogenic potential of a polyherbal formulation Enteropan® was investigated against a multidrug-resistant strain of the bacterium Pseudomonas aeruginosa.
Methods: Growth, pigment production, antibiotic susceptibility, etc., were assessed through appropriate in vitro assays. Virulence of the test pathogen was assessed employing the nematode worm Caenorhabditis elegans as a model host. Molecular mechanisms underlining the anti-pathogenic activity of the test formulation were elucidated through whole transcriptome analysis of the extract-exposed bacterial culture.
Results: Enteropan-pre-exposed P. aeruginosa displayed reduced (~70%↓) virulence towards the model host C. elegans. Enteropan affected various traits like biofilm formation, protein synthesis and secretion, quorum-modulated pigment production, antibiotic susceptibility, nitrogen metabolism, etc., in this pathogen. P. aeruginosa could not develop complete resistance to the virulence-attenuating activity of Enteropan even after repeated exposure to this polyherbal formulation. Whole transcriptome analysis showed 17% of P. aeruginosa genome to get differentially expressed under influence of Enteropan. Major mechanisms through which Enteropan exerted its anti-virulence activity were found to be generation of nitrosative stress, oxidative stress, envelop stress, quorum modulation, disturbance of protein homeostasis and metal homeostasis. Network analysis of the differently expressed genes resulted in identification of 10 proteins with high network centrality as potential targets from among the downregulated genes. Differential expression of genes coding for five (rpoA, tig, rpsB, rpsL, and rpsJ) of these targets was validated through real-time polymerase chain reaction too, and they can further be pursued as potential targets by various drug discovery programmes.
Keywords: Antimicrobial resistance (AMR), Anti-virulence, Caenorhabditis elegans, Iron/Sulphur homeostasis, Metal homeostasis, Network analysis, Nitrosative stress, Polyherbal, Transcriptome
Received: April 4, 2024
Accepted: August 1, 2024
Published online: August 30, 2024
This article includes supplementary material
Corresponding author:
Vijay Kothari
email: vijay.kothari@nirmauni.ac.in
Drug Target Insights - ISSN 1177-3928 - www.aboutscience.eu/dti
© 2024 The Authors. This article is published by AboutScience and licensed under Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0).
Commercial use is not permitted and is subject to Publisher’s permissions. Full information is available at www.aboutscience.eu
Introduction
Antibiotic-resistant strains of the gram-negative bacterial pathogen Pseudomonas aeruginosa are responsible for considerable morbidity and mortality globally (1). As per CDC’s Antibiotic Resistance Threat Report (2019), multidrug-resistant (MDR) P. aeruginosa was responsible for an estimated 32,600 hospitalizations, and 2,700 deaths in 2017 (2). Healthcare costs in the United States alone attributed to these infections were estimated to be 767 million USD. Later released update to this report indicated an increase in number of infections caused by hospital-onset MDR P. aeruginosa in 2022 as compared to that in 2019 (Online). According to a study conducted by Wattal et al (3), India holds a leading position globally with respect to consumption of antibiotics for human use. This heavy antibiotic usage contributes significantly towards antibiotic resistance, resulting in a substantial increase in mortality among newborns who contract sepsis caused by MDR pathogens. P. aeruginosa displays versatility with respect to types of infections it causes, as it has been involved in pneumonia, urinary tract infections, bloodstream infections, and surgical site infections. Hospitalized patients on ventilators, those with catheters, surgical wounds, or burns are particularly at higher risk of contracting P. aeruginosa infection (4). While treating MDR P. aeruginosa infections, the choice of available effective antibiotics remains quite narrow (5), and hence there is an urgent need for discovery and development of novel antibacterial agents/formulations against this notorious pathogen.
Traditional medicine (TM) formulations can be a potential source of novel leads against bacterial pathogens including P. aeruginosa. Since TM often relies on polyherbal formulations (6,7) for treatment, and these polyherbal formulations display ‘multiplicity of targets’ (8) against susceptible pathogens, investigating the effect of such formulations in the pathogens at whole metabolome or transcriptome level can result in discovery of new molecular targets in pathogens. While dearth of novel cellular and molecular targets has been one of the major hurdles in new antibiotic discovery (9,10), elucidating the anti-pathogenic potential of polyherbal formulations at molecular level can be quite relevant. Often these polyherbal formulations exhibit anti-virulence effect (11) rather than directly inhibiting growth of the target pathogens. They may do so by affecting expression of non-essential genes (i.e. other than housekeeping genes) in the pathogens.
Present study investigated one polyherbal formulation (Enteropan) for its anti-pathogenic potential against a MDR P. aeruginosa. This formulation or its component plant extracts are traditionally being prescribed for treatment of irritable bowel syndrome, diarrhoea, dysentery, and other gastrointestinal problems (Online).
Materials and methods
Test formulation
Test formulation Enteropan® was procured from Dr. Palep’s Medical Research Foundation Pvt. Ltd, Mumbai. All the nine ingredient plants and their parts whose hydroalcoholic extracts have been mixed to prepare this formulation are listed in Table 1. We obtained the polyherbal mix from the manufacturer in dried powder form without any bulking agent, and mixed 4 g of it in 8 mL of dimethylsulfoxide (DMSO; Merck). The DMSO-soluble fraction of Enteropan was found to be 71.17% ± 3.07. After separating the insoluble fraction through centrifugation, the remaining DMSO-dissolved fraction was stored under refrigeration.
Test organisms
P. aeruginosa strain was sourced from our internal lab culture collection. Its antibiogram (Tab. S1) generated through disc diffusion assay revealed it to be resistant to three different classes of antibiotics, that is, co-trimoxazole (combination of trimethoprim and sulfamethoxazole), streptomycin (aminoglycoside), and augmentin (combination of amoxicillin and clavulanic acid). Antibiotic susceptibility of this strain to kanamycin was classified as ‘intermediate’. Pseudomonas broth (20 g/L peptic digest of animal tissue, 10 g/L potassium sulphate, 1.4 g/L magnesium chloride, 3% v/v glycerol, pH 7.0 ± 0.2) or agar (HiMedia) was used to cultivate this bacterium. Inoculum density of this bacterium to be used in all experiments was adjusted at OD625 = 0.08-0.10 to achieve equivalence to McFarland turbidity standard 0.5.
| Scientific name | Common English name
(Indian name) |
Part used | Proportion in the polyherbal mix (mg/capsule) |
|---|---|---|---|
| Aegle marmelos | Wood apple
(Bael) |
Leaves | 100 |
| Myristica fragrans | Nutmeg
(Jaiphal) |
Fruit | 15 |
| Zingiber officinale | Ginger
(Sunthi) |
Rhizome | 60 |
| Aconitum heterophyllum | Indian Atees
(Ativisha) |
Bulb | 30 |
| Coriandrum sativum | Coriander
(Dhanyak) |
Seeds | 40 |
| Cyperus rotundus | Nutgrass
(Nagarmotha) |
Rhizome | 25 |
| Vetiveria zizanioides | Khas Khas grass
(Usheer) |
Roots | 50 |
| Punica granatum | Pomegranate
(Dadim) |
Rind | 50 |
| Holarrhena antidysenterica | Star gooseberry
(Kutaj) |
Bark | 27 |
Escherichia coli OP50 procured from LabTIE B.V. the Netherlands was used as food for Caenorhabditis elegans, while maintaining the worm on NGM (nematode growth medium; 3 g/L NaCl, 1 M CaCl2, 1 M MgSO4, 2.5 g/L peptone, 5 mg/mL cholesterol, 1 M phosphate buffer of pH 6, 17 g/L agar-agar).
The nematode worm C. elegans (N2 Bristol, procured from IIT Gandhinagar, Gujarat, India) was used as a model host for P. aeruginosa. The worm was maintained on NGM agar plates. Worm synchronization was done as described in literature (12) and in our previous studies (13,14) too. Prior to all in vivo assays, worms were kept without food for 2 days to make them gnotobiotic.
In vivo assays
Four different types of in vivo assays performed are described below. Each of them involved live-dead counting over a period of 5 days under a microscope (4×) with halogen light source. On the last day of the experiment, when plates could be opened, death was confirmed by touching them with a straight wire, wherein no movement was considered as confirmation of death. In each well, there were 10 worms in M9 buffer (3 g/L KH2PO4; 6 g/L Na2HPO4; 5 g/L NaCl), which were challenged with P. aeruginosa by adding 100 µL (OD764= 1.50 ± 0.05) of bacterial culture grown in Pseudomonas broth for 21 ± 1 hours at 35 ± 0.5°C.
Anti-pathogenic assay
Ten worms of L3-L4 stage contained in 900 µL M9 buffer were challenged with P. aeruginosa (100 µL of the culture broth) in absence or presence of Enteropan (50-1,000 µg/mL), wherein neither the bacterium nor the worms were pre-exposed to Enteropan. Incubation was done at 22°C for 5 days with live-dead microscopic count once a day.
Anti-infective assay
P. aeruginosa was grown at 35°C for 20-22 hours in Pseudomonas broth with or without Enteropan (5-1,000 µg/mL). Post-incubation, 100 µL of the culture broth was mixed with 900 µL of M9 buffer containing 10 worms (L3-L4 stage) in a 24-well plate (surface non-treated; HiMedia). This plate was incubated at 22°C for 5 days (15,16).
Prophylactic assay
Gnotobiotic worms were incubated in M9 buffer supplemented with Enteropan (5-1,000 µg/mL) for 96 hours. Following incubation, these worms were washed with M9 buffer twice, and then challenged with P. aeruginosa not pre-exposed to Enteropan (100 µL of the culture broth) in 24-well plates (HiMedia). Worm survival was monitored over a 5-day period under microscope (17).
Post-infection assay
Ten worms contained in M9 buffer were first challenged with pathogen, and after allowing P. aeruginosa (100 µL of the culture broth) for 3 or 6 hours to establish infection, Enteropan (50-1,000 µg/mL) was added into the well as a possible post-infection therapy. Survival of worms was observed through a live-dead count under microscope over 5 days (18).
Appropriate controls were included in all the above experiments as relevant:
Sterility Control: Sterile M9 buffer containing neither bacteria nor worms
Survival Control: M9 buffer containing 10 worms (no bacteria added)
Toxicity Control: 10 worms in M9 buffer supplemented with Enteropan
Infection Control: 10 worms in M9 buffer + 100 µL of the P. aeruginosa culture broth (OD764 = 1.50 ± 0.05). These wells did not contain any plant extract.
Vehicle Control: 0.5% v/v DMSO was used in place of Enteropan.
Positive Control: Standard antibiotics employed as positive controls are detailed in the figure legends.
In vitro assays
Growth and pigment quantification
The broth dilution assay was used to evaluate P. aeruginosa’s growth and quorum sensing (QS)-regulated pigment synthesis in the presence or absence of the test formulation. Different concentrations (ranging from 5 to 1,000 μg/mL) of Enteropan formulation were used to challenge the organism. The growth media employed was Pseudomonas broth, into which bacterial inoculum set to 0.5 McFarland turbidity standard was added at 10% v/v, followed by incubation at 35oC for 20-22 hours, with intermittent shaking. The experiment also contained an appropriate vehicle control with DMSO (0.5% v/v) and an abiotic control with extract and growth medium but no inoculum.
Bacterial growth was measured photometrically at the end of the incubation by measuring the culture density at 764 nm (Agilent Cary 60 UV-visible spectrophotometer) (19). Following this, pigment was extracted and quantified in accordance with the procedure outlined for each pigment.
One mL of culture broth was mixed in a 2:1 ratio with chloroform (Merck, Mumbai), followed by centrifugation (15,300 g) for 10 minutes. This resulted in the formation of two immiscible layers. OD of the upper aqueous layer containing the yellow-green fluorescent pigment pyoverdine was measured at 405 nm. Pyoverdine Unit was calculated as OD405/OD764. The lower chloroform layer containing the blue pigment pyocyanin was mixed with 0.1 N HCl (20% v/v; Merck). This caused a change of colour from blue to pink. This was followed by centrifugation (15,300 g) for 10 minutes, and OD of upper layer acidic liquid containing pyocyanin was quantified at 520 nm. Pyocyanin Unit was calculated as OD520/OD764.
Biofilm assays
Biofilm formation is an important virulence trait, and hence the effect of Enteropan on biofilm forming ability of P. aeruginosa, as well as on pre-formed biofilm was investigated. A flow diagram depicting all four different biofilm assays is included in supplementary file (Fig. S2). Biofilm quantification was achieved through crystal violet assay (20). Biofilm viability was assessed through 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (21).
For the crystal violet assay, the biofilm-containing tubes (after discarding the inside liquid) were washed with phosphate-buffered saline (PBS) in order to remove all non-adherent (planktonic) bacteria, and air-dried for 15 minutes. Then, each of the washed tubes was stained with 1.5 mL of 0.4% aqueous crystal violet (Central Drug House, Delhi) solution for 30 minutes. Afterwards, each tube was washed twice with 2 mL of sterile distilled water and immediately de-stained with 1.5 mL of 95% ethanol. After 45 minutes of de-staining, 1 mL of de-staining solution was transferred into separate tubes, and read at 580 nm (Agilent Cary 60 UV-Vis).
For the MTT assay, the biofilm-containing tubes (after discarding the inside liquid) were washed with PBS in order to remove all non-adherent (planktonic) bacteria, and air-dried for 15 minutes. Then 1.8 mL of minimal media (sucrose 15 g/L, K2HPO4 5 g/L, NH4Cl 2 g/L, NaCl 1 g/L, MgSO4 0.1 g/L, yeast extract 0.1 g/L, pH 7.4 ± 0.2) was added into each tube, followed by addition of 200 μL of 0.3% MTT [HiMedia]. Then after 2 hours of incubation at 35°C, all liquid content was discarded, and the remaining purple formazan derivatives were dissolved in 2 mL of DMSO and measured at 540 nm.
Nitrite estimation
Quantification of nitrite in bacterial culture was achieved through a colorimetric assay using modified Griess reagent (22,23). Supernatant (250 µL) obtained from centrifugation (13,500 g; 25°C; 10 minutes) of P. aeruginosa culture grown in the presence or absence of Enteropan was mixed with 250 µL of Griess reagent (1×; Sigma-Aldrich), followed by 15 minutes of incubation in dark at room temperature. Absorbance of the resulting pink colour was measured at 540 nm. This absorbance was plotted on the standard curve prepared using NaNO2 (0.43-65 µM) to calculate nitrite concentration. Sodium nitroprusside (Sigma-Aldrich) was used as a positive control, as it is known to generate nitrosative stress in bacteria (24). Deionized water was used as negative control.
Antibiotic susceptibility test
Antibiogram of P. aeruginosa’s overnight grown culture in Pseudomonas broth in the presence or absence of Enteropan was generated through disc diffusion assay in accordance with Clinical and Laboratory Standards Institute (CLSI) guidelines (25). Cells grown in Pseudomonas broth were separated through centrifugation (13,600 g) and washed with phosphate buffer (pH 7.0 ± 0.2) followed by centrifugation. The resulting cell pellet was used to prepare inoculum for subsequent disc diffusion assay by suspending the cells in normal saline and adjusting the OD625 between 0.08 and 0.10. This inoculum (100 µL) was spread onto cation-adjusted Mueller-Hinton agar (HiMedia) plates (Borosil; 150 mm) followed by placing the antibiotic discs (Icosa G-I MINUS; HiMedia, Mumbai) on the agar surface. Incubation at 35°C was made for 18 ± 1 hours, followed by observation and measurement of zone of inhibition.
Protein estimation
Extracellular protein present in bacterial culture (grown in the presence or absence of Enteropan) supernatant, and intracellular protein in the cell lysate was quantified through Folin-Lowry method (26,27). After measuring cell density, 1 mL of P. aeruginosa culture was centrifuged (13,600 g), and the resulting supernatant was used for extracellular protein estimation. The remaining cell pellet was subjected to lysis (28) for release of intracellular proteins. Briefly, the cell pellet was washed with phosphate buffer (pH 7.4) and centrifuged (13,600 g). Resulting pellet was resuspended in 1 mL of chilled lysis buffer (0.876 g NaCl, 1 mL of Triton X-100, 0.5 g sodium deoxycholate, 0.1 g sodium dodecyl sulphate, and 0.60 g Tris HCl, in 99 mL of distilled water) and centrifuged (500 rpm) for 30 minutes at 4°C for agitation purpose. This was followed by further centrifugation (16,000 g at 4°C) for 20 min. Resulting cell lysate (supernatant) was used for protein estimation. Kanamycin (HiMedia; at IC50: 200 µg/mL), an aminoglycoside antibiotic known to inhibit bacterial protein synthesis (29,30), was used as a positive control.
Whole transcriptome analysis
To gain insight into the molecular mechanisms by which Enteropan attenuates bacterial virulence and modulates various traits like pigment production, antibiotic susceptibility, nitrogen metabolism, protein synthesis/excretion, etc., we compared the gene expression profile of Enteropan-pre-treated P. aeruginosa with that of control culture at the whole transcriptome level.
RNA extraction
Trizol (Invitrogen Bioservices; 343909) was used to extract RNA from bacterial cells (31). RNA was dissolved in nuclease-free water after precipitation with isopropanol and washing with 75% ethanol. Using the RNA HS assay kit (Thermo Fisher; Q32851) and adhering to the manufacturer’s instructions, the extracted RNA was quantified using a Qubit 4.0 fluorometer (Thermo Fisher; Q33238). RNA concentration and purity were evaluated using Nanodrop 1000. Finally, RNA was checked on the TapeStation using HS RNA ScreenTape (Agilent) to yield RIN (RNA Integrity Number) values (Tab. S2).
Library preparation
Final libraries were measured using a Qubit 4.0 fluorometer (Thermo Fisher; Q33238), a DNA HS assay kit (Thermo Fisher; Q322851), and a TapeStation 4150 (Agilent) using high-sensitivity D1000 ScreenTapes (Agilent; 5067-5582). The acquired sizes of all libraries are reported in Table S3.
Genome annotation and functional analysis
FastQC v.0.11.9 (default parameters) was used to undertake a quality assessment of the sample’s raw fastq readings (32). The reads’ quality was then reevaluated using Fastq v.0.20.1 (33) after pre-processing the raw fastq reads with Fastq v.0.20.1.
The P. aeruginosa genome (GCA_000006765.1_ASM676v1) was indexed using bowtie2-build (34) v2.4.2 (default parameters). The processed reads were mapped to the P. aeruginosa genome using bowtie2 v2.4.2. Gene counts were determined using feature count v.0.46.1 (35) to quantify the aligned reads from the individual samples. Differential expression was estimated using the exact test (parameters: dispersion 0.1) with these gene counts as inputs in edgeR (36). The up- and downregulated sequences were extracted from the P. aeruginosa coding file and annotated using Blast2GO (37) to obtain the Gene Ontology (GO) keywords. These GO terms were used to create GO bar graphs with the wego tool (38).
All the raw sequence data has been submitted to the Sequence Read Archive. The relevant accession number is SRX15248092 (Online).
Network analysis
Network analysis was carried out for Enteropan-exposed P. aeruginosa’s differentially expressed genes (DEG) fulfilling the dual criteria of log fold change ≥2 and false discovery rate (FDR) ≤0.001. List of DEG was fed into the database STRING (v.11.5) (39) for generating the Protein-Protein Interaction (PPI) network. Then the genes were arranged in decreasing order of ‘node degree’ (a measure of connectivity with other genes or proteins), and those above an empirically selected threshold value (19 and 53 for up- and downregulated genes, respectively) were subjected to ranking by cytoHubba (v.3.9.1) plugin (40) of Cytoscape (41). Since cytoHubba uses 12 different ranking methods, we considered the DEG being top-ranked by ≥6 different methods (i.e. 50% of the total ranking methods) for further analysis. These top-ranked shortlisted proteins were further subjected to network cluster analysis through STRING, and those that were part of multiple clusters were considered as ‘hubs’ which can be taken up for further confirmation of their targetability. Here ‘hub’ refers to a gene or protein interacting with many other genes/proteins. Hubs thus identified were further subjected to co-occurrence analysis to see whether an anti-virulence agent targeting them is likely to satisfy the criterion of selective toxicity (i.e. targeting the pathogen without harming host). This sequence analysis allowed us to end with a limited number of proteins which satisfied various statistical and biological significance criteria simultaneously, that is, (1) log fold change ≥2; (2) FDR ≤0.001; (3) relative higher node degree; (4) top-ranking by at least six cytoHubba methods; (5) member of more than one local network cluster; (6) high probability of the target being absent from the host.
Real-time polymerase chain reaction analysis
Polymerase chain reaction (PCR) was used to confirm the differential expression of the possible hubs discovered by network analysis of the DEG reported from whole transcriptome analysis (WTA). Primer3Plus (42) was used to design primers for the target genes (Tab. 2). These primer sequences were verified for their ability to specifically bind only to the target gene sequence throughout the whole genome file of P. aeruginosa. RNA extraction and purity check was executed as described in the previous section. The SuperScript™ VILO™ cDNA Synthesis Kit (Invitrogen Biosciences) was used to generate complementary DNA. Using gene-specific primers purchased from Sigma-Aldrich, the PCR experiment was carried out employing the temperature profile shown in Table S4. The gene PA3725 (recJ) was kept as an endogenous control. The reaction mix used was FastStart Essential DNA Green Master mix (Roche; 06402712001). Real-time PCR (RT-PCR) assay was performed on QuantStudio 5 RT-PCR machine (Thermo Fisher Scientific, USA). Sample generation for PCR validation was done independent of that for transcriptome assay.
| Gene ID | Primers | Amplicon size (bp) |
|---|---|---|
| PA4238 (rpoA) | FP: 5′-CGCTGAACATGAAGCTGAAG-3′
RP: 5′-CAGGACCAGTTTGTCCAGGT-3′ |
194 |
| PA1800 (tig) | FP: 5′-ACCGAAGTCAACAAGCGTCT-3′
RP: 5′-GGATTCAGCTTCTGCTCGAC-3′ |
208 |
| PA3656 (rpsB) | FP: 5′-GCGCAACAAGATCCATATCA-3′
RP: 5′-GATCGACTGACGGATGGTCT-3′ |
232 |
| PA4268 (rpsL) | FP: 5′-TACATCGGTGGTGAAGGTCA-3′
RP: 5′-TACTTCGAACGACCCTGCTT-3′ |
155 |
| PA4264 (rpsJ) | FP: 5′-GATTCGGTTGAAGGCTTTTG-3′
RP: 5′-TACTGATCACGCGCATCTTT-3′ |
174 |
| Control gene
PA3725 (recJ) |
FP: 5′-CCAGTTGAGCATCCAGGAGT-3′
RP: 5′-TTTCAGCACCAGCTTCAGGT-3′ |
157 |
Statistical analysis
All results reported are means of three or more independent experiments, each performed in triplicate. Statistical significance was assessed through t-test performed using Microsoft Excel®, and data with p ≤ 0.05 was considered to be significant.
Results and discussion
In vivo assays
P. aeruginosa displayed reduced virulence towards C. elegans in the presence of Enteropan
When C. elegans was challenged with P. aeruginosa in the presence of Enteropan, the bacterium could kill lesser worms than in the absence of Enteropan (Fig. 1A; Supplementary videos: A-E). The most effective concentration of Enteropan with respect to offering protection to the worm population from bacterial attack was found to be 250 µg/mL. Since higher concentrations offered either at par or lesser protection to the worms, the dose-response relationship here can be said to be nonlinear. Irrespective of the magnitude of protection offered to worm population in the presence of Enteropan®, progeny worms were observed (third day onwards) in all experimental and positive control wells, but not in the wells pertaining to vehicle control. It might have occurred that the virulence-attenuated P. aeruginosa were used by the worms as food, and that allowed them to reproduce.
Enteropan pre-treatment reduced bacterial virulence towards C. elegans
Enteropan (5-1,000 μg/mL)-pre-treated P. aeruginosa was found to exert lesser virulence against C. elegans than its not-exposed counterpart. Enteropan concentrations ≤250 µg/mL could not compromise P. aeruginosa’s ability to kill C. elegans; however, concentrations ≥500 µg/mL did compromise bacterial virulence significantly (Fig. 1B; Supplementary videos: F-I). Enteropan pre-treatment of the pathogen at these effective concentrations not only attenuated bacterial virulence but also supported worm fertility as evidenced by the appearance of numerous progenies in the experimental wells by the fifth day. The most effective concentration of Enteropan with respect to virulence-attenuating effect was found to be 600 µg/mL, as the effect of higher concentrations till 1 mg/mL was statistically not different than that of 600 µg/mL.
After confirming the anti-virulence activity of Enteropan against P. aeruginosa, we asked whether this pathogen can develop resistance upon repeatedly getting exposed to the test formulation. To answer this, we subcultured P. aeruginosa in Pseudomonas broth supplemented with Enteropan (600 µg/mL) multiple times. Enteropan-pre-exposed P. aeruginosa thus obtained after fifth and tenth such subculturing were allowed to attack C. elegans in M9 buffer (containing no Enteropan). No resistance seemed to have evolved in P. aeruginosa till fifth subculturing; however, 10 subculturings in Enteropan-supplemented media seemed to allow the pathogen to overcome this formulation’s anti-virulence effect marginally (23.3%; Fig. 1C). This inability of the pathogen to develop complete resistance against Enteropan might be attributable to the polyherbal nature of the formulation. As the polyherbal formulations can have multiple bioactive compounds in them, they may exert a multiplicity of targets against the susceptible pathogen. To develop resistance in this scenario, the pathogen would be required to develop multiple simultaneous mutations, and that is a quite less probable event biologically as well statistically.
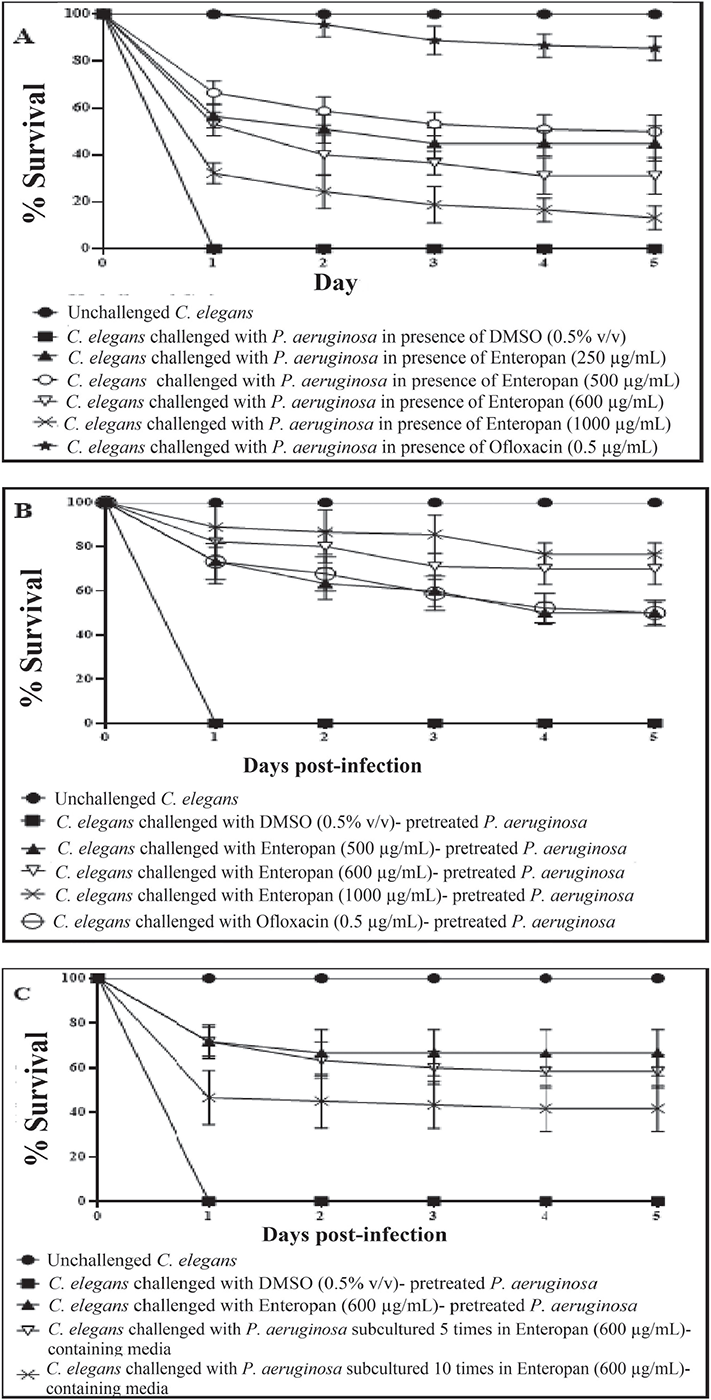
FIGURE 1 - Enteropan attenuates Pseudomonas aeruginosa’s virulence towards the model host Caenorhabditis elegans. Dimethylsulfoxide (DMSO) present in the ‘vehicle control’ at 0.5% v/v did not affect bacterial virulence. Neither DMSO nor Enteropan showed any toxicity towards the worm population at tested concentrations. To avoid overcrowding in the figures (A, B), we have not shown lines corresponding to concentrations which had no effect on bacterial virulence, and also that for 750 μg/mL, as its virulence-attenuating effect was statistically a part of that of 600 μg/mL. Supplementary videos pertaining to these experiments are available at: osf.io/fnywk. All the % values reported in this figure legend are statistically significant at p<0.001.
A) P. aeruginosa’s virulence towards the host worm gets attenuated in the presence of Enteropan. Enteropan conferred a survival benefit on host C. elegans at concentrations of 250, 500, 600, 750, and 1,000 μg/mL with survival rates of 45% ± 5.47, 50% ± 7.07, 31.11% ± 7.8, 35% ± 8.36, and 13.33% ± 5, respectively. Ofloxacin (0.5 µg/mL) was employed as a positive control and conferred 85.55% ± 5.27 survival benefit on host worm. Progenies (TNTC – too numerous to count) were observed on the third day in experimental wells and positive control. See supplementary videos A-E. B) Enteropan pre-treatment reduced bacterial virulence towards C. elegans. Pre-treatment of bacteria with Enteropan at concentrations of 500, 600, 750, and 1000 μg/mL reduced its virulence towards host worm by 50% ± 5, 70% ± 7.07, 78.88% ± 6, and 76.66% ± 5, respectively, as per the fifth day end-point. Ofloxacin (0.5 µg/mL) pre-treatment reduced bacterial virulence towards the host worm by 50% ± 5.77. Progenies were observed on the third day in experimental wells corresponding to ≥500 μg/mL Enteropan as well as positive control. See supplementary videos F-I. C) P. aeruginosa did not develop complete resistance even after repeated exposure to Enteropan. P. aeruginosa obtained after fifth and tenth subculturings in Enteropan (600 μg/mL)-containing media displayed 58.33%±4.04 and 43.33%±10.3 lesser virulence, respectively, than extract-non-exposed pathogen.
Enteropan offered prophylactic protection to C. elegans
To investigate whether Enteropan pre-feeding can offer any prophylactic benefit to worm population in the face of subsequent pathogen challenge, we allowed P. aeruginosa to attack worms pre-fed with Enteropan (5-1,000 µg/mL). Enteropan at ≥50 µg/mL did confer prophylactic benefit on worm population. While concentrations till 500 µg/mL supported worm survival (13%-26%) till 48 hours post-pathogen challenge (Fig. 2A), higher concentrations supported worm survival (20%-23%) till the fifth day (Fig. 2B; Supplementary videos: J-M). Considering the first-day end-point (i.e. by the time the pathogen killed 100% worms in control wells), certain Enteropan concentrations (500-750 µg/mL) performed at par to the positive control ofloxacin.
Enteropan is effective as a post-infection therapy
To investigate whether Enteropan is effective as a post-infection therapy, we added Enteropan (50-1,000 μg/mL) after 3 or 6 hours of mixing bacteria with the worms. While Enteropan addition post 6 hours of bacterial attack on worms could not rescue the host (Fig. S1), its addition post 3 hours of bacterial attack could rescue 10%-27% of the worms (Fig. 2C; Supplementary videos: N-R). As a post-infection therapy, Enteropan’s performance did not improve with increase in concentration.
To have a mechanistic insight into the Enteropan-P. aeruginosa interaction, we checked the effect of Enteropan on various virulence traits of this pathogen in vitro, and also compared the gene expression profile of the Enteropan-treated P. aeruginosa with that of extract-non-exposed control at the whole transcriptome level.
In vitro experiments
Enteropan forced overproduction of QS-regulated pigments without affecting the bacterial growth heavily
Enteropan till 100 µg/mL had no effect on P. aeruginosa growth. Though from 250 µg/mL onwards it had some growth inhibitory effect, the magnitude of this inhibitory effect did not increase much with increase in concentration (Fig. 3A). Except 5 µg/mL, Enteropan at all tested concentrations enhanced production of QS-regulated pigments (pyoverdine and pyocyanin). Hence concentrations of 25-100 µg/mL can be said to have pure quorum-modulatory effect on P. aeruginosa. Though in general Enteropan’s effect on pigment production seemed to be dose-dependent, bacterium’s response at 600 µg/mL seemed to deviate from this pattern in case of both the pigments.
Enteropan had a moderately negative effect on pre-formed biofilm
Though Enteropan’s presence could not compromise P. aeruginosa’s ability to form biofilm, when Enteropan was added onto the pre-formed biofilm, it could eradicate the biofilm partly and also reduced the metabolic activity within the biofilm (Fig. 3B).
Enteropan disturbed nitrogen metabolism in P. aeruginosa
Since multiple genes associated with detoxification of reactive nitrogen species were upregulated (transcriptome data described later), we hypothesized that Enteropan-treated P. aeruginosa’s ability to overcome nitrosative stress is compromised. To check this hypothesis, we quantified nitrite concentration in extract-treated P. aeruginosa culture supernatant, wherein it was found to have 47.10% higher nitrite concentration as compared to control (Fig. 3C). This higher accumulation of nitrite can be taken as an indication of compromised denitrification efficiency, since nitrite is an important intermediate in denitrification pathway ahead of the toxic nitric oxide (43). Nitrosative stress can impact the overall bacterial fitness negatively in multiple ways (44). Reactive nitrogen species can damage biomolecules like DNA, lipids, and proteins. Resistance to nitrosative stresses is of crucial importance towards the survival of bacteria in the environment as well as inside the host. In gram-negative bacteria, several mechanisms protecting against oxidative and nitrosative stresses are present in the envelope. Excessive nitrosative stress can disturb the envelope homeostasis, and this in fact is reflected in the transcriptome of Enteropan-exposed P. aeruginosa, wherein 32 cell envelope (cell wall and lipopolysaccharide, LPS)-associated genes are expressed differently.
Enteropan-modulated P. aeruginosa’s susceptibility to imipenem and augmentin
When Enteropan-pre-treated P. aeruginosa cells were subsequently challenged with different antibiotics in a disc diffusion assay, these cells exhibited marginal increase in their susceptibility to imipenem; however, their susceptibility to augmentin disappeared following Enteropan pre-treatment (Tab. S5; Fig. 3D, E). This effect of Enteropan pre-treatment on imipenem susceptibility was also confirmed in liquid culture, wherein Enteropan-pre-treated cells were observed to exhibit up to 21.43% higher susceptibility to imipenem (Fig. 3F). Imipenem belongs to the carbapenem class of beta-lactams (45), and carbapenem resistance among P. aeruginosa isolates are being viewed as a serious problem (46). Since this class of antibiotics are looked as a last resort for treatment of MDR P. aeruginosa (47), resistance modifiers capable of making this bacterium more susceptible to them can be of help in extending the lifespan of these antibiotics by allowing their use at lower doses. However as seen with augmentin in this study, the effect of herbals on antibiotic susceptibility of pathogen may not always be favourable.
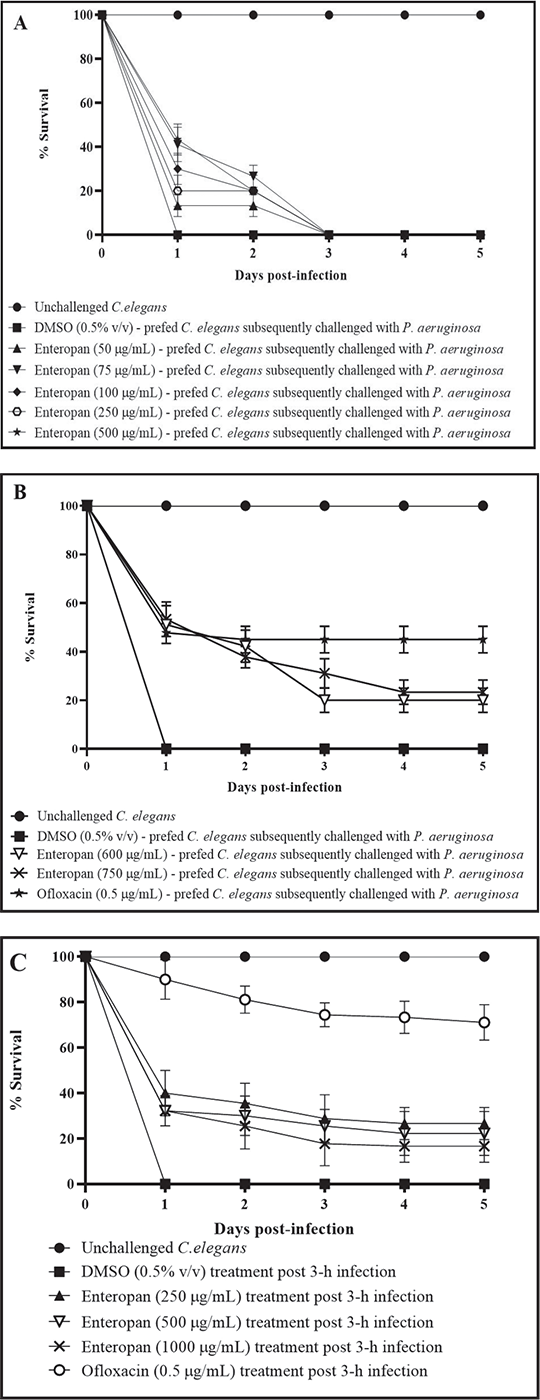
FIGURE 2 - Prophylactic and post-infection therapeutic potential of Enteropan. All the % values reported in this figure legend are statistically significant at p<0.001. A-B) Enteropan offered prophylactic protection to the worm population against subsequent bacterial challenge. Worms pre-fed with Enteropan concentrations of 50, 75, 100, 250, and 500 μg/mL registered 13.3% ±5, 26.6% ±5, 20% ±8.66, 17% ± 6.66, and 20% ±8.66 better survival, respectively, till the second day in the face of subsequent pathogen challenge. Worms pre-fed with higher concentrations of Enteropan at 600, 750, and 1000 μg/mL registered 51.11% ± 7.8, 53.33 ± 7.07, and 43.33% ± 8.66 survival, respectively, till the end of first day of pathogen challenge. Magnitude of the prophylactic benefit as per fifth day point was 20% ± 5, 23.33 ± 5, and 20% ± 7.07, respectively. Pre-feeding the worms with dimethylsulfoxide (DMSO, 0.5% v/v) did not alter their susceptibility to subsequent bacterial challenge. Ofloxacin (0.5 µg/mL) employed as a positive control conferred 44.9% ± 5.49 prophylactic benefit on the host worms. See supplementary videos J-M. C) Enteropan could partially rescue worm population when used as a post-infection therapy. When pre-infected worms were exposed to Enteropan at 250, 500, 600, 750, and 1000 μg/mL, they scored 26.66% ± 7.0, 22.22% ± 9.71, 11.1% ± 3.33, 13.3% ± 6, and 16.66% ± 7.07 better survival, respectively, than control worms. Ofloxacin (0.5 µg/mL) employed as positive control, 3 hours post-infection, rescued 71.11% ± 7.81 worms. DMSO (0.5% v/v) did not confer any survival benefit when added post-infection. See supplementary videos N-R.
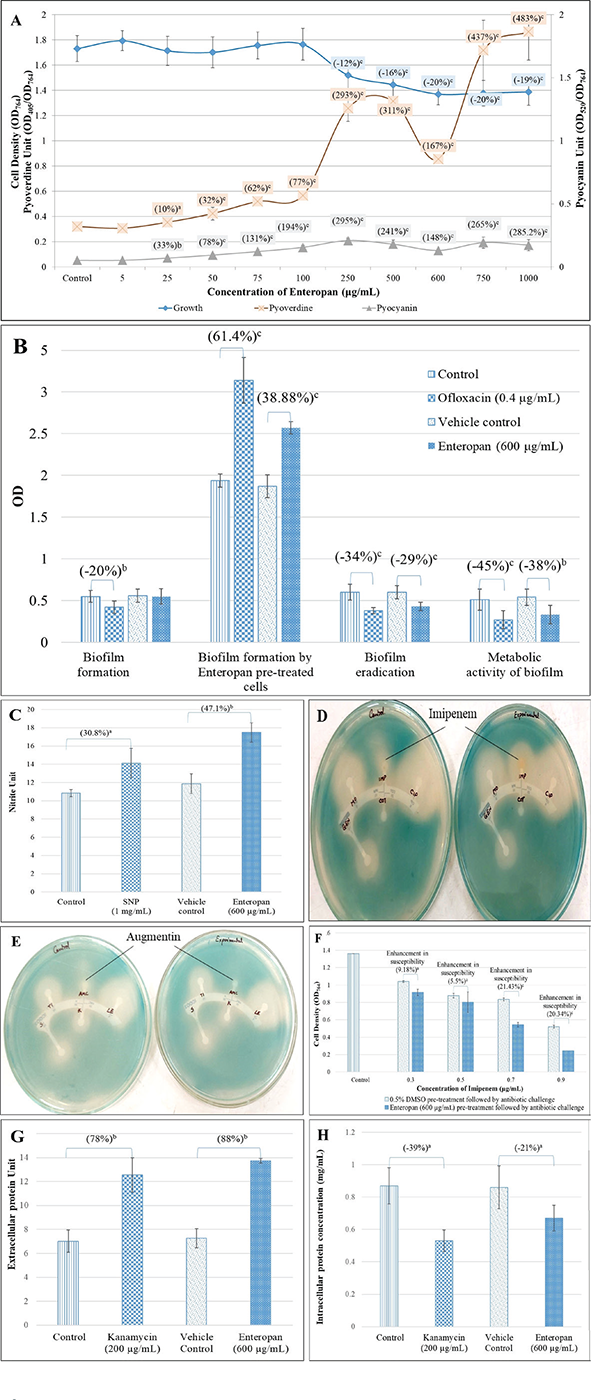
FIGURE 3 - Enteropan’s effect on various phenotypic traits of Pseudomonas aeruginosa revealed through different in vitro assays. A) Enteropan enhances production of quorum-regulated pigments in P. aeruginosa, while exhibiting a mild growth inhibitory effect. Bacterial growth was measured as OD764. Pyoverdine Unit and Pyocyanin Unit were calculated as the ratio OD405/OD764 and OD520/OD764 (an indication of pyoverdine and pyocyanin production per unit of growth), respectively; ‘Control’ shown in this figure is the vehicle control (0.5% v/v dimethylsulfoxide (DMSO)), which affected neither growth nor pigment production. Ofloxacin (0.5 µg/mL) inhibited growth by 65.6% ±5.32, while inhibiting pigment production completely. B) Enteropan’s effect on P. aeruginosa’s biofilm formation capability and on pre-formed biofilm. While P. aeruginosa’s biofilm formation ability remained unaffected in the presence of Enteropan, Enteropan-pre-exposed cells subsequently allowed to form biofilm on glass surface accumulated higher biomass. Enteropan when added onto pre-formed biofilm could eradicate the biofilm partially, and also reduced the biofilm metabolic activity notably. DMSO (0.5% v/v) used as vehicle control did not affect biofilm of the P. aeruginosa in any of these four assays. C) P. aeruginosa culture accumulated higher extracellular nitrite in the presence of Enteropan. While nitrite concentration in vehicle control (P. aeruginosa supplemented with 0.5% v/v DMSO) was at par to that without DMSO, Enteropan caused nitrite concentration in P. aeruginosa culture supernatant to rise. Sodium nitroprusside used as positive control caused 30.8% higher nitrite build-up in P. aeruginosa culture. Nitrite Unit (i.e. Nitrite concentration:cell density ratio was calculated to nullify any effect of cell density on nitrite production). D, E) Enteropan-pretreated cells responded to certain antibiotics differently. Enteropan-pre-exposed P. aeruginosa experienced an increased or decreased susceptibility to imipenem and augmentin, respectively, as revealed in disc diffusion assay. F) Enteropan pre-treatment enhanced P. aeruginosa’s susceptibility to imipenem, as revealed in the broth dilution assay. G) Increased extracellular protein content in P. aeruginosa culture grown in the presence of Enteropan. Protein Unit was calculated as ratio of OD750/OD764 (an indication of protein production per unit of growth). H) Reduced intracellular protein content in P. aeruginosa grown in the presence of Enteropan. Protein content reported in mg/mL are cell density neutralized values, wherein OD764 was adjusted to 1.00 prior to cell lysis. Kanamycin employed as a positive control at its sub-minimum inhibitory concentration level also generated response similar to that of Enteropan from bacterial culture with respect to extracellular and intracellular protein content. DMSO (0.5% v/v) used as ‘vehicle control’ affected neither extracellular nor intracellular protein content. ap ≤ 0.05, bp ≤ 0.01, cp ≤ 0.001.
Enteropan alters protein synthesis and secretion in P. aeruginosa
Extracellular protein concentration (after nullifying cell density) in P. aeruginosa culture supernatant in the presence of Enteropan was found to be 1.89-fold higher than that in the absence of Enteropan (Fig. 3G). Cell density-neutralized intracellular protein concentration of P. aeruginosa cells grown in the presence of Enteropan was found to be 1.40-fold lower than cells grown in the absence of Enteropan (Fig. 3H). It seems that Enteropan exerted an inhibitory effect on protein synthesis in P. aeruginosa and promoted protein export. This might have caused even some of the essential proteins to leave the cell. The increased export of proteins by Enteropan-treated cells may be assumed to have originated from overexpression of efflux pump/transport machinery (as suggested by the transcriptome data too described later) and a compromised cell envelope integrity suggested by differential expression of 32 genes involved in cell wall or LPS synthesis. Kanamycin, a known inhibitor of protein synthesis in bacteria, was employed as a positive control in this assay. Kanamycin belongs to the aminoglycoside group of antibiotics, which at sub-minimum inhibitory concentration (MIC) level caused P. aeruginosa culture supernatants to have 1.79-fold higher extracellular protein. Such increase in extracellular protein concentration in P. aeruginosa exposed to sub-MIC level of kanamycin was also reported by Takahashi et al (48).
Enteropan treatment causes large-scale differential gene expression in P. aeruginosa
A whole transcriptome level comparison of the gene expression profile of Enteropan (600 µg/mL)-treated P. aeruginosa with that of control revealed a total of 952 genes getting expressed differentially (log fold change ≥2 and FDR ≤0.001). This amounted to differential expression of 17% of genome, wherein 616 genes were upregulated (Tab. S6) and 336 were downregulated (Tab. S9). Corresponding volcano plot (Fig. S3) is given in supplementary file. A function-wise categorization of all the DEG is presented in Figure 4. While all DEG pertaining to cell division were downregulated (and majority of DEG pertaining to translation too), the majority of DEG associated with efflux pump/transport were upregulated. Overexpression of efflux machinery is known to compromise bacterial fitness by causing physiological dysregulation (49). Owing to the important physiological roles of efflux pumps in various functions such as intercellular communication, bacterial pathogenicity and virulence, and biofilm formation, expression of majority of them is subject to tight control by different transcriptional regulators. Any mischief with this regulation leading to overexpression of the efflux function may result in leaking of even essential items. An empirical look at the list of DEG suggested that Enteropan attenuated virulence of P. aeruginosa by causing dysregulation of metal homeostasis, nitrogen metabolism, transcription, amino acid and protein synthesis, carbon metabolism, motility, efflux, etc. Results of various in vitro assays presented in the preceding section corroborates well with the transcriptome data.
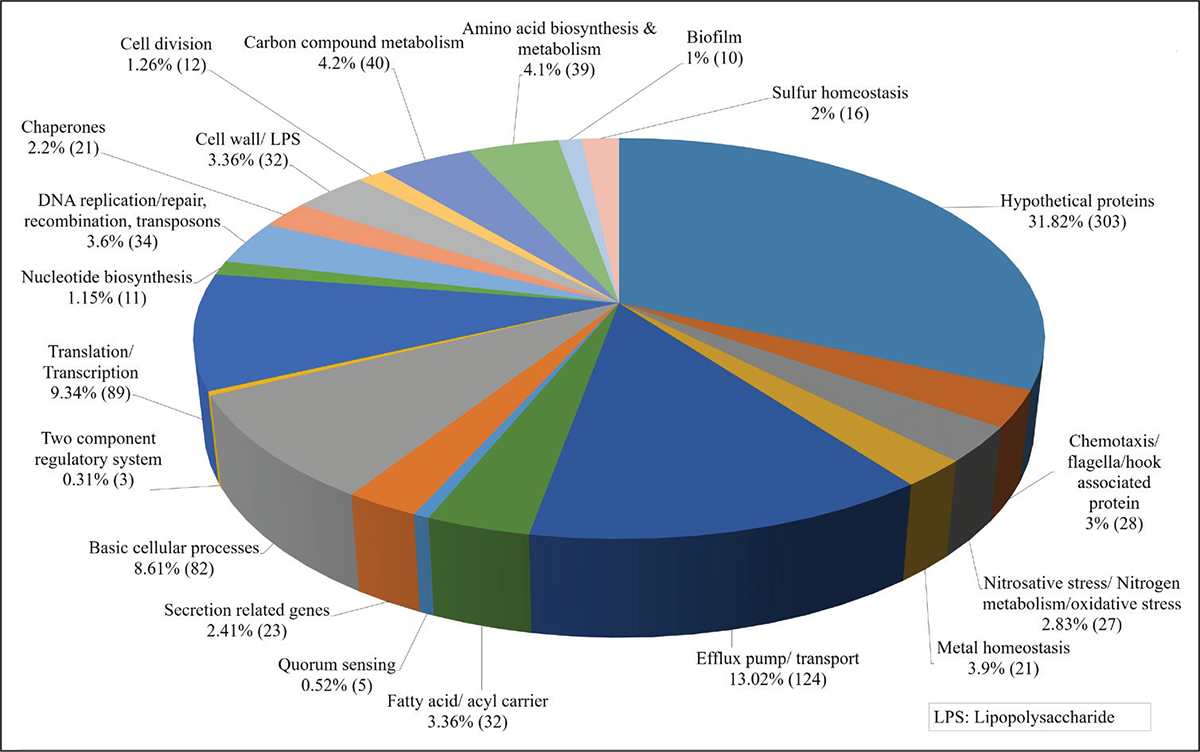
FIGURE 4 - Function-wise categorization of the differentially expressed genes (DEG) in Enteropan-treated Pseudomonas aeruginosa. Percent values reported are calculated considering the total number of differently expressed genes as 100%. Values in parenthesis are number of DEG belonging to that particular category.
Network analysis of DEG in Enteropan-exposed P. aeruginosa
We created PPI network for up- and downregulated genes separately. PPI network for upregulated genes generated through STRING is presented in Figure 5A, which shows 610 nodes connected through 2,272 edges with an average node degree of 7.45. Since the number of edges in this PPI network is 2.06-fold higher than expected (1,101) with a PPI enrichment p-value <1.0e-16, this network can be said to possess significantly more interactions among the member proteins that what can be expected for a random set of proteins of the identical sample size and degree distribution. Such enrichment can be taken as an indication of the member proteins being at least partially biologically connected. When we arranged all the upregulated DEGs in decreasing order of node degree, 572 nodes were found to have a non-zero score (Tab. S7), and we selected top 52 genes with a node degree ≥19 for further ranking by different cytoHubba methods. Then we looked for genes which appeared among top-6 ranked candidates by ≥6 cytoHubba methods (Tab. S8), and six such genes were further checked for interactions among themselves by cluster analysis (Fig. 5B), whose overexpression can be hypothesized to disturb pathogen physiology. Interaction map of these six potential hubs showed them to be strongly networked as the average node degree score was 5. This network possessed 15 edges as against expected (zero) for any such random set of proteins. The PPI network showed five of these six potential hubs to be part of a single local network cluster (Fig. 5B). Co-occurrence analysis showed all of these six hubs being absent from humans (Tab. 3) and hence agonists of these hubs may be expected to target pathogen selectively without interfering with host system functioning.
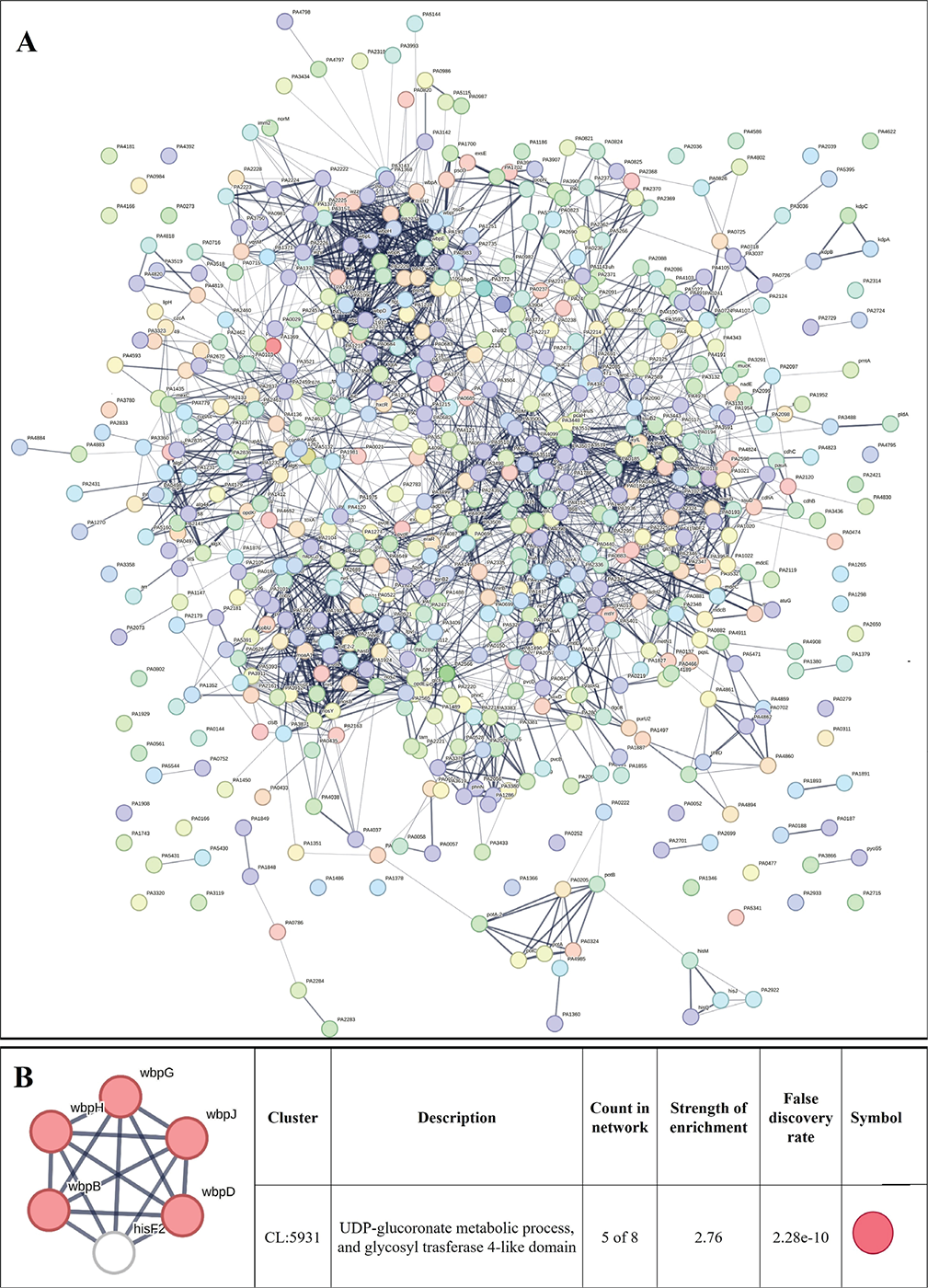
FIGURE 5 - A) Protein-Protein Interaction (PPI) network of upregulated genes in Enteropan-exposed Pseudomonas aeruginosa. Edges represent protein-protein associations that are meant to be specific and meaningful, that is, proteins jointly contribute to a shared function. This does not necessarily mean they are physically binding to each other. Network nodes represent all the proteins produced by a single, protein-coding gene locus. B) PPI network of top-ranked genes revealed through cytoHubba among upregulated differentially expressed genes (DEG) in Enteropan-exposed P. aeruginosa.
PPI network for downregulated genes is presented in Figure 6A, which shows 327 nodes connected through 3,206 edges with an average node degree of 19.6. Since the number of edges in this PPI network is 1.93-fold higher than expected (1,658) with a PPI enrichment p-value <1.0e-16, this network can be said to possess significantly more interactions among the member proteins that what can be expected for a random set of proteins of the identical sample size and degree distribution. Such enrichment can be taken as an indication of the member proteins being at least partially biologically connected. When we arranged all the downregulated DEGs in decreasing order of node degree, 298 nodes were found to have a non-zero score, and we selected top 50 genes with a node degree ≥53 (Tab. S10) for further ranking by different cytoHubba methods. Then we looked for genes which appear among top-10 ranked candidates by ≥6 cytoHubba methods, and 10 such genes (Tab. S11) were identified as potential hubs, whose downregulation can be hypothesized to attenuate P. aeruginosa virulence. Interaction map of these 10 potential hubs (Fig. 6B) showed them to be strongly networked as the average node degree score was 9. This network possessed 45 edges as against expected (15) for any such random set of proteins. The PPI network showed these 10 genes to be distributed among six different local network clusters, whose strength score ranged from 1.97 to 2.2 (Fig. 6B). Co-occurrence analysis (Tab. 3) of these 10 hub proteins indicated four (rpsD, rpsJ, rpoA, rplF) of them to be absent from humans, and hence they can be said to possess high targetability with respect to discovery of new antibiotics satisfying the criteria of selective toxicity. Since all the 10 predicted hubs are indicated by co-occurrence analysis to be present in other important bacterial pathogens too, antagonists of these proteins are likely to be useful as broad-spectrum antibiotics.
TABLE 3 - Co-occurrence analysis of genes coding for potential targets in P. aeruginosa

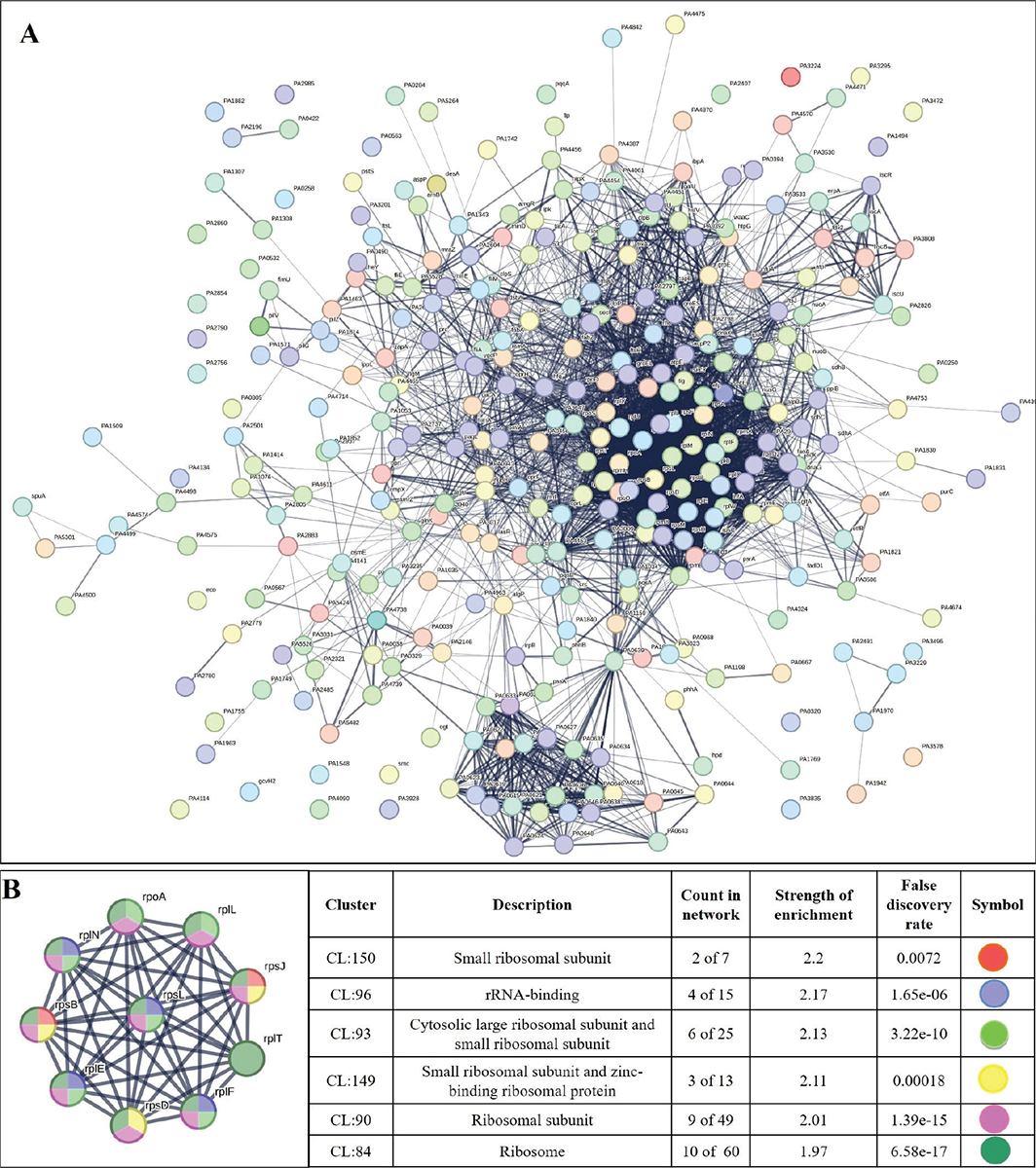
FIGURE 6 - A) Protein-Protein Interaction (PPI) network of downregulated genes in Enteropan-exposed Pseudomonas aeruginosa. B) PPI network of top-ranked genes revealed through cytoHubba among downregulated differentially expressed genes (DEG) in Enteropan-exposed P. aeruginosa.
Target validation through RT-PCR
From the 10 identified hubs among the downregulated genes, we selected five (rpoA, tig, rpsB, rpsL, rpsJ) for further validation through RT-PCR. From the 10 genes shown in Figure 6B, four (rpoA, rpsB, rpsL, rpsJ) passing the dual criteria of node degree ≥70 (Tab. S10) and been part of ≥3 clusters were selected for RT-PCR. Though rpsD (node degree 70) also passed these dual criteria, since already three rps genes were selected for PCR, we preferred rpoA (node degree 75) over it. Additionally, we included one gene (tig; 3.73-fold↓; node degree 70) for PCR validation, though it was not among the identified hubs, because tig is a trigger factor involved in protein export, and we did observe a heavy increase in extracellular protein content in Enteropan-exposed P. aeruginosa. PCR did confirm downregulation of all the selected five genes in Enteropan-exposed P. aeruginosa (Fig. 7), and thus they can be considered as potential antibacterial targets worthy of attention by drug discovery programmes.
Conclusion
The polyherbal formulation Enteropan was found to have virulence-attenuating effect against an important gram-negative bacterial pathogen P. aeruginosa, without affecting its growth heavily. As can be expected from any multicomponent polyherbal formulation, Enteropan also exerted multiplicity of targets against the test pathogen. A large fraction of the bacterial genome was expressed differently under influence of this anti-pathogenic formulation, which corroborated well with the altered phenotypic traits in extract-exposed bacterial culture. Major mechanisms revealed various in vitro/in vivo assays and transcriptome analysis through which Enteropan exerted its anti-virulence activity were found to be generating nitrosative stress, oxidative stress, quorum modulation, disturbance of protein homeostasis, and metal homeostasis. A wholistic summary depicting the mechanistic details associated with the anti-pathogenic potential of Enteropan against P. aeruginosa is presented in Figure 8, with particular attention on Enteropan’s effect on QS machinery and virulence regulators of this notorious pathogen. Our results validate the anti-pathogenic potential of Enteropan, and also the concept of polyherbalism and its relevance in combating antimicrobial resistance.
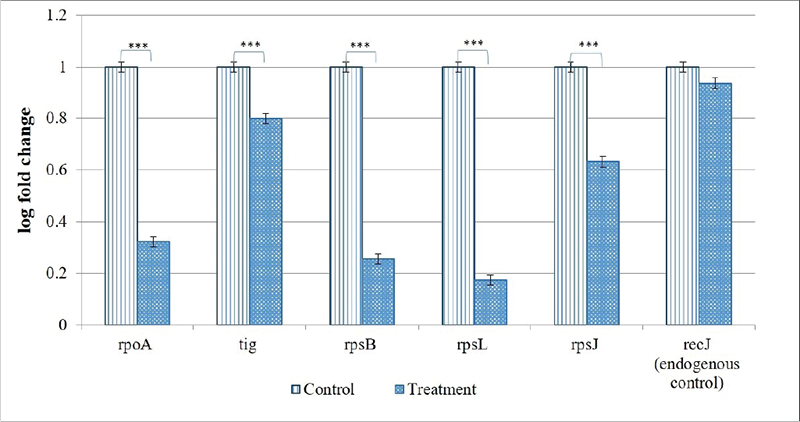
FIGURE 7 - Confirmation of differential expression of selected genes in Enteropan-treated Pseudomonas aeruginosa through real-time polymerase chain reaction (RT-PCR). recJ selected as an endogenous control was not expressed differently (false discovery rate 1) between control and experimental bacterial cultures. ***p ≤ 0.001.
Acknowledgements
The authors thank Nirma Education and Research Foundation (NERF), Ahmedabad, for infrastructural support; Virupakshi Soppina (IIT-Gn) for providing worms. SP and GG acknowledge fellowship from Gujarat government under their SHODH scheme. NT acknowledges fellowship from Nirma University.
Disclosures
Conflict of interest: HSP, is involved in manufacturing and marketing of the formulation Enteropan®; however, that in no way has affected the design of experiments or interpretation of results. Other authors declare no competing interest.
Financial support: This study did not receive any extramural financial support.
Data availability statement: All the relevant data has been included in the main manuscript and supplementary files.
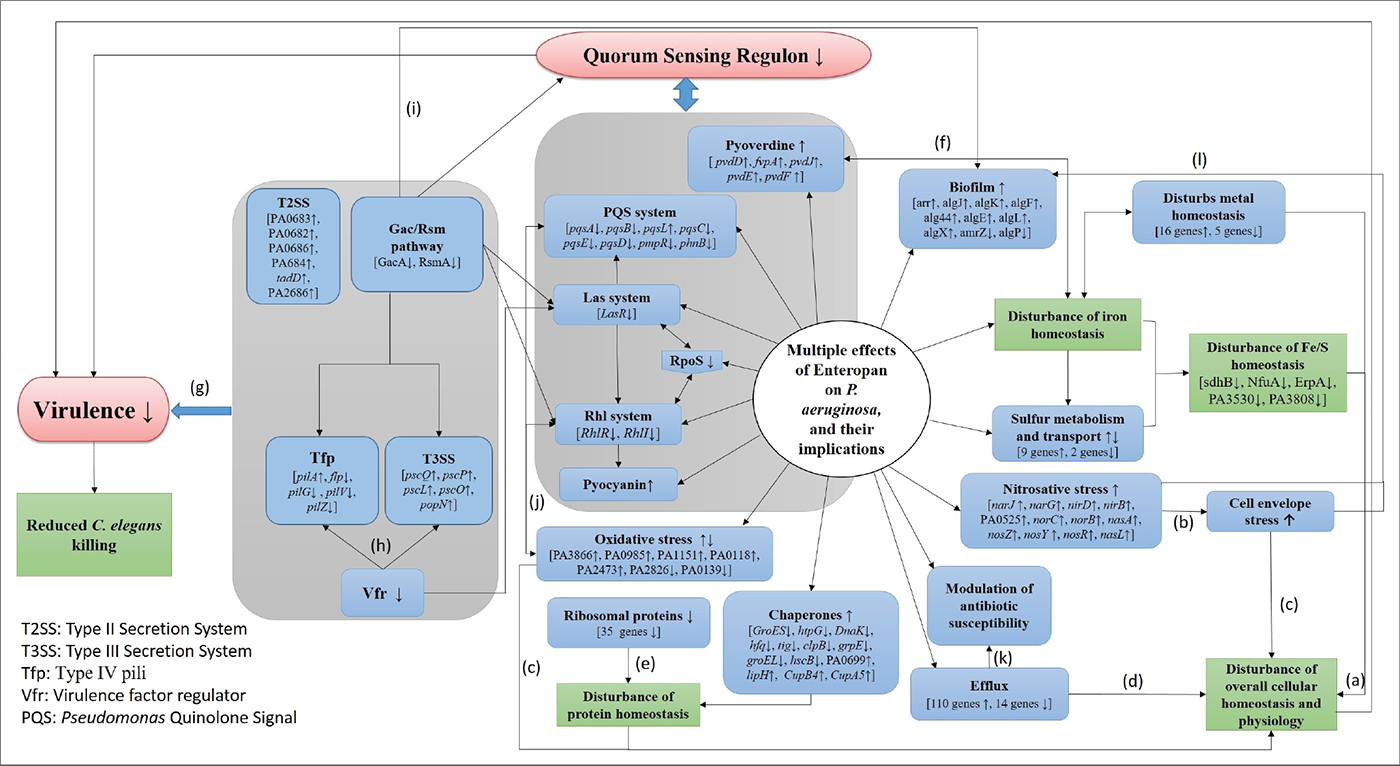
FIGURE 8 - Large-scale disturbance of transcriptional regulation in Pseudomonas aeruginosa caused by Enteropan compromises its overall cellular homeostasis and virulence. This figure presents a wholistic summary of multiple effects exerted by Enteropan against P. aeruginosa. Various cellular, physiological, and virulence-associated traits of P. aeruginosa expressed differently under the influence of Enteropan are depicted. The genes shown with an up or down arrow are those getting differentially expressed with a log fold change of ≥1.5 and false discovery rate (FDR) ≤0.05. The Gac/Rsm pathway inversely regulates the expression of virulence factors (T3SS, Tfp, exopolysaccharides) associated with acute and chronic disease (50). T2SS is responsible for secreting many secretory proteins like alkaline phosphatase, lipase, exotoxin A, phospholipase, and proteases. T3SS is largely involved in secretion of virulence determinants associated with acute infection. Vfr is a global regulator of virulence gene expression, which allows coordinated production of related virulence functions (Tfp, T3SS) necessary for adherence to an intoxication of host cells. LasR is a transcriptional activator of multiple virulence-associated genes in P. aeruginosa. It represents a central checkpoint, with the highest degree of interconnection in the network. RpoS, the stationary phase sigma factor, influences the expression of more than one-third of all the quorum-regulated genes. It is a central regulator of many stationary phase-inducible genes and a master stress-response regulator under a variety of stress conditions (51). Tfp, a major surface adhesin, mechanochemically regulates virulence factors in P. aeruginosa (52). The Rhl system is a quorum sensing system acquired by P. aeruginosa through lateral gene transfer. PQS is an essential mediator of the shaping of the population structure of P. aeruginosa and of its response to and survival in stress conditions. a(53); b(44); c(54); d(55); e(56); f(57); g(58); h(59); i(60), j(61, 62), k(63); l(64).
References
- 1. Kunz Coyne AJ, El Ghali A, Holger D, Rebold N, Rybak MJ. Therapeutic strategies for emerging multidrug-resistant Pseudomonas aeruginosa. Infect Dis Ther. 2022;11(2):661-682. CrossRef PubMed
- 2. Craig M. CDC’s Antibiotic Resistance Threats Report 2019. Extended spectrum β-lactamase (ESBL)-producing Enterobacteriaceae. CDC; 2019.
- 3. Wattal C, Kler N, Oberoi JK, Fursule A, Kumar A, Thakur A. Neonatal sepsis: mortality and morbidity in neonatal sepsis due to multidrug-resistant (MDR) organisms: part 1. Indian J Pediatr. 2020;87(2):117-121. CrossRef PubMed
- 4. Sendra E, Fernández-Muñoz A, Zamorano L, et al. Impact of multidrug resistance on the virulence and fitness of Pseudomonas aeruginosa: a microbiological and clinical perspective. Infection. 2024 Aug;52(4):1235-1268. CrossRef PubMed.
- 5. Diggle SP, Whiteley M. Microbe profile: Pseudomonas aeruginosa: opportunistic pathogen and lab rat. Microbiology (Reading). 2020;166(1):30-33. CrossRef PubMed
- 6. Parasuraman S, Thing GS, Dhanaraj SA. Polyherbal formulation: concept of Ayurveda. Pharmacogn Rev. 2014;8(16):73-80. CrossRef PubMed
- 7. Benamar-Aissa B, Gourine N, Ouinten M, Yousfi M. Synergistic effects of essential oils and phenolic extracts on antimicrobial activities using blends of Artemisia campestris, Artemisia herba alba, and Citrus aurantium. Biomol Concepts. 2024 Feb 14;15(1). CrossRef PubMed
- 8. Makhoba XH, Viegas C Jr, Mosa RA, Viegas FPD, Pooe OJ. Potential impact of the multi-target drug approach in the treatment of some complex diseases. Drug Des Devel Ther. 2020;14:3235-3249. CrossRef PubMed
- 9. Kumar M, Sarma DK, Shubham S, et al. Futuristic non-antibiotic therapies to combat antibiotic resistance: a review. Front Microbiol. 2021;12:609459. CrossRef PubMed
- 10. Ruparel FJ, Shah SK, Patel JH, Thakkar NR, Gajera GN, Kothari VO. Network analysis for identifying potential anti-virulence targets from whole transcriptome of Pseudomonas aeruginosa and Staphylococcus aureus exposed to certain anti-pathogenic polyherbal formulations. Drug Target Insights. 2023;17(1):58-69. CrossRef PubMed
- 11. Allen RC, Popat R, Diggle SP, Brown SP. Targeting virulence: can we make evolution-proof drugs? Nat Rev Microbiol. 2014;12(4):300-308. CrossRef PubMed
- 12. Corsi AK, Wightman B, Chalfie M. A transparent window into biology: a primer on Caenorhabditis elegans. Genetics. 2015;200(2):387-407. CrossRef PubMed
- 13. Patel H, Patel F, Jani V, et al. Anti-pathogenic potential of a classical Ayurvedic Triphala formulation. F1000Res. 2019 Jul 18;8:1126. CrossRef. PubMed;
- 14. Joshi C, Patel P, Palep H, Kothari V. Validation of the anti-infective potential of a polyherbal ‘Panchvalkal’ preparation, and elucidation of the molecular basis underlining its efficacy against Pseudomonas aeruginosa. BMC Complement Altern Med. 2019;19(1):19. CrossRef PubMed
- 15. Durai S, Vigneshwari L, Balamurugan K. Caenorhabditis elegans-based in vivo screening of bioactives from marine sponge-associated bacteria against Vibrio alginolyticus. J Appl Microbiol. 2013;115(6):1329-1342. CrossRef PubMed
- 16. Patel P, Joshi C, Palep H, Kothari V. Anti-infective potential of a quorum modulatory polyherbal extract (panchvalkal) against certain pathogenic bacteria. J Ayurveda Integr Med. 2020;11(3):336-343. CrossRef PubMed
- 17. Patel P, Joshi C, Funde S, Palep H, Kothari V. Prophylactic potential of a Panchgavya formulation against certain pathogenic bacteria. F1000Res. 2018 Oct 8;7:1612. CrossRef. PubMed
- 18. Patel P, Joshi C, Kothari V. Anti-pathogenic efficacy and molecular targets of a polyherbal wound- care formulation (Herboheal) against Staphylococcus aureus. Infect Disord Drug Targets. 2019;19(2):193-206. CrossRef. PubMed.
- 19. Joshi C, Kothari V, Patel P. Importance of selecting appropriate wavelength, while quantifying growth and production of quorum sensing regulated pigments in bacteria. Recent Pat Biotechnol. 2016;10(2):145-152. CrossRef PubMed
- 20. Hirshfield IN, Barua S, Basu P. Overview of biofilms and some key methods for their study. In: Goldman E, Green LH, eds. Practical handbook of microbiology. 2nd ed. CRC Press; 2008:695-708:chap 42.
- 21. Trafny EA, Lewandowski R, Zawistowska-Marciniak I, Stępińska M. Use of MTT assay for determination of the biofilm formation capacity of microorganisms in metalworking fluids. World J Microbiol Biotechnol. 2013;29(9):1635-1643. CrossRef PubMed
- 22. Misko TP, Schilling RJ, Salvemini D, Moore WM, Currie MG. A fluorometric assay for the measurement of nitrite in biological samples. Anal Biochem. 1993;214(1):11-16. CrossRef PubMed
- 23. Yildirim K, Atas C, Tanyel Akcit E, et al. Nitrate reductase assay for rapid determination of methicillin-resistant Staphylococcus aureus clinical isolates. Lab Med. 2024;55(2):174-178. CrossRef CrossRef PubMed
- 24. Barnes RJ, Bandi RR, Wong WS, et al. Optimal dosing regimen of nitric oxide donor compounds for the reduction of Pseudomonas aeruginosa biofilm and isolates from wastewater membranes. Biofouling. 2013;29(2):203-212. CrossRef PubMed
- 25. Simner PJ, Hindler JA, Bhowmick T, et al. What’s new in antibiograms? Updating CLSI M39 guidance with current trends. J Clin Microbiol. 2022;60(10):e0221021. CrossRef PubMed
- 26. Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193(1):265-275. CrossRef PubMed
- 27. Dulley JR, Grieve PA. A simple technique for eliminating interference by detergents in the Lowry method of protein determination. Anal Biochem. 1975;64(1):136-141. CrossRef PubMed
- 28. Mishra M, Tiwari S, Gomes AV. Protein purification and analysis: next generation Western blotting techniques. Expert Rev Proteomics. 2017;14(11):1037-1053. CrossRef PubMed
- 29. Suzuki J, Kunimoto T, Hori M. Effects of kanamycin on protein synthesis: inhibition of elongation of peptide chains. J Antibiot (Tokyo). 1970;23(2):99-101. CrossRef PubMed
- 30. Ullah H, Ali S. Classification of anti-bacterial agents and their functions. Antibacterial Agents. 2017;10:1-6. CrossRef
- 31. Jahn CE, Charkowski AO, Willis DK. Evaluation of isolation methods and RNA integrity for bacterial RNA quantitation. J Microbiol Methods. 2008;75(2):318-324. CrossRef PubMed
- 32. Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. Online
- 33. Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018 Sep 1;34(17):i884-i890. CrossRef. PubMed
- 34. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357-359. CrossRef PubMed
- 35. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923-930. CrossRef PubMed
- 36. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139-140. CrossRef PubMed
- 37. Conesa A, Götz S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics. 2008;2008:619832. CrossRef PubMed
- 38. Ye J, Zhang Y, Cui H, et al. WEGO 2.0: a web tool for analyzing and plotting GO annotations, 2018 update. Nucleic Acids Res. 2018;46(W1):W71-W75. CrossRef PubMed
- 39. Szklarczyk D, Kirsch R, Koutrouli M, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51(D1):D638-D646. CrossRef PubMed
- 40. Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(suppl 4):S11. CrossRef PubMed
- 41. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498-2504. Online CrossRef PubMed
- 42. Untergasser A, Cutcutache I, Koressaar T, et al. Primer3 – new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):e115. CrossRef PubMed
- 43. Borrero-de Acuña JM, Timmis KN, Jahn M, Jahn D. Protein complex formation during denitrification by Pseudomonas aeruginosa. Microb Biotechnol. 2017;10(6):1523-1534. CrossRef PubMed
- 44. Chautrand T, Depayras S, Souak D, et al. Gaseous NO2 induces various envelope alterations in Pseudomonas fluorescens MFAF76a. Sci Rep. 2022;12(1):8528. CrossRef PubMed
- 45. Mansour H, Ouweini AEL, Chahine EB, Karaoui LR. Imipenem/cilastatin/relebactam: A new carbapenem β-lactamase inhibitor combination. Am J Health Syst Pharm. 2021;78(8):674-683. CrossRef PubMed
- 46. Souza GHA, Rossato L, Brito GT, Bet GMDS, Simionatto S. Carbapenem-resistant Pseudomonas aeruginosa strains: a worrying health problem in intensive care units. Rev Inst Med Trop Sao Paulo. 2021 Sep 27;63:e71. CrossRef PubMed
- 47. Pragasam AK, Raghanivedha M, Anandan S, Veeraraghavan B. Characterization of Pseudomonas aeruginosa with discrepant carbapenem susceptibility profile. Ann Clin Microbiol Antimicrob. 2016;15(1):12. CrossRef PubMed
- 48. Takahashi E, Lee JM, Mon H, et al. Effect of antibiotics on extracellular protein level in Pseudomonas aeruginosa. Plasmid. 2016;84-85:44-50. CrossRef PubMed
- 49. Sun J, Deng Z, Yan A. Bacterial multidrug efflux pumps: mechanisms, physiology and pharmacological exploitations. Biochem Biophys Res Commun. 2014;453(2):254-267. CrossRef PubMed
- 50. Sultan M, Arya R, Kim KK. Roles of two-component systems in Pseudomonas aeruginosa virulence. Int J Mol Sci. 2021;22(22):12152. CrossRef PubMed
- 51. Murakami K, Ono T, Viducic D, et al. Role for rpoS gene of Pseudomonas aeruginosa in antibiotic tolerance. FEMS Microbiol Lett. 2005;242(1):161-167. CrossRef PubMed
- 52. Persat A, Inclan YF, Engel JN, Stone HA, Gitai Z. Type IV pili mechanochemically regulate virulence factors in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2015;112(24):7563-7568. CrossRef PubMed
- 53. Myriam P, Braulio P, Javiera RA, et al. Insights into systems for iron-sulfur cluster biosynthesis in acidophilic microorganisms. J Microbiol Biotechnol. 2022;32(9):1110-1119. CrossRef PubMed
- 54. Mitchell AM, Silhavy TJ. Envelope stress responses: balancing damage repair and toxicity. Nat Rev Microbiol. 2019;17(7):417-428. CrossRef PubMed
- 55. Hajiagha MN, Kafil HS. Efflux pumps and microbial biofilm formation. Infect Genet Evol. 2023;112:105459. CrossRef PubMed
- 56. Deuerling E, Gamerdinger M, Kreft SG. Chaperone interactions at the ribosome. Cold Spring Harb Perspect Biol. 2019;11(11):a033977. CrossRef PubMed
- 57. Hamza EH, El-Shawadfy AM, Allam AA, Hassanein WA. Study on pyoverdine and biofilm production with detection of LasR gene in MDR Pseudomonas aeruginosa. Saudi J Biol Sci. 2023;30(1):103492. CrossRef PubMed
- 58. Coggan KA, Wolfgang MC. Global regulatory pathways and cross-talk control Pseudomonas aeruginosa environmental lifestyle and virulence phenotype. Curr Issues Mol Biol. 2012;14(2):47-70. PubMed
- 59. Marsden AE, Intile PJ, Schulmeyer KH, et al. Vfr directly activates exsA transcription to regulate expression of the Pseudomonas aeruginosa type III secretion system. J Bacteriol. 2016;198(9):1442-1450. CrossRef PubMed
- 60. Schuster M, Greenberg EP. Regulatory networks in pathogenic bacteria: lessons from cell‐cell communication in Pseudomonas aeruginosa. Virulence mechanisms of bacterial pathogens. ASM Press; 2007:75-88. CrossRef
- 61. Häussler S, Becker T. The pseudomonas quinolone signal (PQS) balances life and death in Pseudomonas aeruginosa populations. PLoS Pathog. 2008;4(9):e1000166. CrossRef PubMed
- 62. Bradley R, Simon D, Spiga L, Xiang Y, Takats Z, Williams H. Laser desorption rapid evaporative ionization mass spectrometry (LD-REIMS) demonstrates a direct impact of hypochlorous acid stress on PQS-mediated quorum sensing in Pseudomonas aeruginosa. mSystems. 2024;9(4):e0116523. CrossRef PubMed
- 63. Xu Y, Zheng X, Zeng W, et al. Mechanisms of heteroresistance and resistance to imipenem in Pseudomonas aeruginosa. Infect Drug Resist. 2020;13:1419-1428. CrossRef PubMed
- 64. Fazeli-Nasab B, Sayyed RZ, Mojahed LS, et al. Biofilm production: a strategic mechanism for survival of microbes under stress conditions. Biocatal Agric Biotechnol. 2022;42:102337. CrossRef