 |
Drug Target Insights 2024; 18: 20-26 ISSN 1177-3928 | DOI: 10.33393/dti.2024.3059 ORIGINAL RESEARCH ARTICLE |
 |
RBD mutations at the residues K417, E484, N501 reduced immunoreactivity with antisera from vaccinated and COVID-19 recovered patients
ABSTRACT
Introduction: It is unclear whether induced spike protein-specific antibodies due to infections with SARS-CoV-2 or to the prototypic Wuhan isolate-based vaccination can immune-react with the emerging variants of SARS-CoV-2.
Aim/objectives: The main objective of the study was to measure the immunoreactivity of induced antibodies postvaccination with Covishield™ (ChAdOx1 nCoV-19 coronavirus vaccines) or infections with SARS-CoV-2 by using selected peptides of the spike protein of wild type and variants of SARS-CoV-2.
Methodology: Thirty patients who had recovered from SARS-CoV-2 infections and 30 individuals vaccinated with both doses of Covishield™ were recruited for the study. Venous blood samples (5 mL) were collected at a single time point from patients within 3-4 weeks of recovery from SARS-CoV-2 infections or receiving both doses of Covishield™ vaccines. The serum levels of total immunoglobulin were measured in both study groups. A total of 12 peptides of 10 to 24 amino acids length spanning to the receptor-binding domain (RBD) of wild type of SARS-CoV-2 and their variants were synthesized. The serum levels of immune-reactive antibodies were measured using these peptides.
Results: The serum levels of total antibodies were found to be significantly (p<0.001) higher in the vaccinated individuals as compared to COVID-19 recovered patients. Our study reported that the mutations in the RBD at the residues K417, E484, and N501 have been associated with reduced immunoreactivity with anti-sera of vaccinated people and COVID-19 recovered patients.
Conclusion: The amino acid substitutions at the RBD of SARS-CoV-2 have been associated with a higher potential to escape the humoral immune response.
Keywords: Antibodies, COVID-19, Immune escape, Immunoreactivity, Mutation, SARS-CoV-2
Received: March 1, 2024
Accepted: May 7, 2024
Published online: May 31, 2024
This article includes supplementary material
Corresponding author:
Dablu Lal Gupta
email: dr.dlgupta@aiimsraipur.edu.in
Drug Target Insights - ISSN 1177-3928 - www.aboutscience.eu/dti
© 2024 The Authors. This article is published by AboutScience and licensed under Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0).
Commercial use is not permitted and is subject to Publisher’s permissions. Full information is available at www.aboutscience.eu
Introduction
The coronavirus disease 2019 (COVID-19) pandemic caused significant global impact and continues to be a threat despite the introduction of vaccines (1,2). The vaccines have been highly effective in preventing COVID-19 around the world (3). Globally, the COVID-19 pandemic is still producing a significant amount of illness and mortality (4). According to current knowledge, mildly symptomatic people represent around 80% of all instances of COVID-19, which can range in severity from asymptomatic to deadly pneumonitis (5). The virus can cause a range of symptoms, from mild to severe, and can be transmitted through the respiratory tract (6). It has significant impacts on at-risk patients and the healthcare system, including high costs, loss of human resources, and psychological problems (7). COVID-19, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), enters human cells by binding the receptor-binding domain (RBD) of its spike (S) protein to the angiotensin-converting enzyme 2 (ACE2) receptor (8,9). Antibodies directed against the S-protein have the potential to neutralize the virus efficiently (10). Since the first case of SARS-CoV-2 in Wuhan, China, over 774 million infections have been documented, leading to the emergence of seven major lineages with various variants (11,12). Coronaviruses have a large RNA genome and mutations can arise naturally during replication, leading to evolutionary advantages such as increased infectivity or improved receptor binding (13). The Omicron variant of SARS-CoV-2 has 30 non-synonymous amino acid mutations in the spike protein, which have been reported to enable immune evasion and reduce vaccine effectiveness (14). Variants of concern (VOCs), such as B.1.1.7, B.1.1.298, B.1.429, P.2, P.1, and B.1.351, have mutations that can increase transmissibility, virulence, or evade immune responses (15,16). These variants mainly have mutations in the spike protein, particularly in the S1 and RBD regions, which are the main targets of neutralizing antibodies.
Twelve vaccines have been approved for use in humans since the first reported case of SARS-CoV-2 in December 2019, with high efficacy in preventing COVID-19 (17). However, the emergence of new variants has raised concerns about the effectiveness of these vaccines, with some trials showing reduced efficacy against variant strains in certain countries (18). The emergence of VOCs with mutations in the S-protein raises concerns about immune evasion. Some VOCs, such as B.1.1.7 and B.1.351, have shown reduced susceptibility to neutralizing antibodies, but the immunoreactivity of induced antibodies after vaccination with individual’s mutations occurred in the RBD in VOCs is not well understood (15,19). The study focuses on understanding the humoral immune responses to SARS-CoV-2 in COVID-19 convalescent donors and vaccine recipients. This study investigated the levels of SARS-CoV-2 spike protein-specific total antibodies in a cohort of individuals who have received both doses of Covishield™ vaccine (vaccinated group) and patients who have recovered from SARS-CoV-2 infections (recovered group). The aim of the study was to compare the immunoreactivity of peptides of wild-type SARS-CoV-2 and peptides of mutated SARS-CoV-2 with anti-sera of vaccinated people and COVID-19 recovered patients to understand the effectiveness of vaccination and potential immune evasion strategies.
Materials and methods
Sample size and study groups
A total of 60 individuals of age ≥18 years (30 patients recovered from COVID-19 infections, 30 received both doses of Covishield™ vaccine) were recruited in the present study. COVID-19 recovered patients of both sexes who had not received any COVID-19 vaccines were recruited for the COVID-19 recovered group, while age-matched individuals without previous history of SARS-CoV-2 infections who have received both doses of Covishield™ were recruited for the vaccinated group. Patients on immunotherapy or having any kind of organ transplantations or immunocompromised patients (patients with human immunodeficiency virus [HIV] infection, autoimmunity, chronic kidney disease [CKD], and any other immunodeficiency diseases) were excluded from the study. This study was approved by the Institute Ethics Committee (IEC), All India Institute of Medical Sciences, Raipur, Chhattisgarh, India (1936/IEC-AIIMSRPR/2021). The completely filled up and signed written consent form was obtained from each participant before taking the blood sample for the study.
Disease severity
Patients who tested positive for SARS-CoV-2 infection by reverse transcriptase-polymerase chain reaction (RT-PCR) and got admitted to the Institute from October 2021 to December 2022 were included in the study. COVID-19 recovered patients were divided into severe and mild groups: severe cases were defined as those requiring invasive mechanical ventilation or high-flow nasal oxygen, and mild cases as neither requiring oxygen nor in-patient hospital care.
Sample collection
A total of 5 mL whole blood sample was collected in a plain vial by venipuncture within 3 to 4 weeks of recovery from COVID-19 infections, while the same amount of blood sample was collected within 3 to 4 weeks postvaccination with both doses of Covishield™. The serum was collected to measure the levels of total antibodies and immunoreactivity of antibodies with selected peptides of spike protein of wild type and variants of SARS-CoV-2.
Serum isolation
The blood samples collected in the plain vial were kept for 30 minutes for clotting. After that, it was centrifuged at 1,500 rpm for 10 minutes at 4°C. The resulting supernatant called serum was separated into fresh Eppendorf tubes and stored at −80°C till further use.
Determination of serum levels of total antibodies
The serum levels of total antibodies (immunoglobulin, Ig) were measured in the collected serum samples of the vaccinated individuals and COVID-19 recovered patients. Human SARS-CoV-2 spike (trimer) Ig total enzyme-linked immunosorbent assay (ELISA) kit (Invitrogen, ThermoFisher Scientific) was used for the estimation of total immunoglobulins as per recommended standard protocol. Ten healthy serum samples collected before 2019 were used as controls to investigate the specificity and sensitivity of ELISA kit.
Peptide synthesis and purification
A total of 12 peptides (six for the wild-type Wuhan isolates and six for the SARS-CoV-2 variants) of RBD of the spike protein were selected based on the available sequence of SARS-CoV-2 through UniPort software. These peptides were commercially synthesized and obtained to test the antibody responses. The peptides P1, P3, P5, P7, P9, and P11 matched with wild-type SARS-CoV-2 sequence, while peptides P2, P4, P6, P8, P10, and P12 matched with variants of SARS-CoV-2 (Tab. 1). The peptides P1 and P2 represent wild-type and mutated SARS-CoV-2, respectively. Both peptides P1 and P2 have same number of amino acids and same sequences of amino acids (411-424), but differ from each other at single amino acid residue at position 417. Likewise, peptides P3 and P4 have same amino acid sequence and length (470-483), but differ from each other at two residues at positions 477 and 478 in wild-type and mutated SARS-CoV-2, respectively. The peptides P5 and P6 show wild-type and mutated SARS-CoV-2, respectively, at residue 484. Both peptides have a single amino acid difference at residue 484. Similarly, peptides P7, P9, P11 of wild-type SARS-CoV-2 and peptides P8, P10, and P12 of SARS-CoV-2 variants have same length and sequence of amino acids, but differ from one another by one or more than one amino acid (Tab. 1).
| S. No | Peptides | Sequences of amino acids in the RBD of SARS-CoV-2 | Sequence number | Mutating sites | VOC/wild type |
|---|---|---|---|---|---|
| 1 | P1 | APGQTGKIADYNYK | 411-424 | K417 | Wild |
| 2 | P2 | APGQTGNIADYNYK | 411-424 | 417N | Common site of mutation for Delta and Omicron |
| 3 | P3 | TEIYQAGSTPCNGV | 470-483 | S477, T478 | Wild |
| 4 | P4 | TEIYQAGNKPCNGV | 470-483 | 477N, 478K | Common site of mutation for Delta and Omicron |
| 5 | P5 | PCNGVEGFNCYFPL | 479-492 | E484 | Wild |
| 6 | P6 | PCNGVAGFNCYFPL | 479-492 | 484A | Common site of mutation for Delta and Omicron |
| 7 | P7 | PLQSYGFQPTNGVG | 491-504 | N501 | Wild |
| 8 | P8 | PLQSYGFQPTYGVG | 491-504 | 501Y | Common site of mutation for Delta and Omicron |
| 9 | P9 | WNSNNLDSKVSGNYN | 436-450 | N440, G446 | Wild |
| 10 | P10 | WNSNKLDSKVSGNYN | 436-450 | 440K, 446S | Omicron |
| 11 | P11 | YFPLQSYGFQPTNGVGYQPYR | 489-509 | Q493, G496, Q498, N501, Y505 | Wild |
| 12 | P12 | YFPLRSYSFRPTYGVGHQPYR | 489-509 | 493R, 496S, 498R, 501Y, 505H | Omicron |
RBD = receptor-binding domain; VOC = variants of concern.
The sites of amino acid variations in the peptide sequences are highlighted in bold.
Direct binding assay
To check the immunoreactivity of peptides with SARS-CoV-2 anti-sera and vaccine anti-sera, direct binding assay was performed using standard ELISA protocol. The ELISA plate was coated with individual peptides (200 ng/well) of spike protein. After blocking and washing SARS-CoV-2 anti-sera and vaccine anti-sera were added at a dilution of 1:200 onto the ELISA plate and incubated at 37°C for 2 hours. After washing, anti-human IgG antibody conjugated with horse radish peroxidase (HRP; 1:200 dilutions) was added and incubated for 1 hour at 37°C. Washing step was repeated after incubation and 100 μL/well substrate solution (TMB) was added. The color was developed within 10-15 minutes. The color reaction was stopped by adding 50 μL/well of 2N H2SO4. The plate was read at 450 nm in the ELISA reader.
Statistical analysis
Data visualization was performed via GraphPad Prism version 8.0 (GraphPad software, San Diego, CA, USA) and STATA 12 (STATA software, Chicago, USA) for Windows 10.0. Result was expressed as mean and standard deviation (SD) for normal distribution, or median and interquartile range for skewed distribution. For two-group analysis, we used the Mann-Whitney U-test for continuous variables. The level of significance (p-value) in the mean antibody titer was compared and analyzed with the Student’s t-test.
Results
Clinical and demographic details
The study analyzed 30 COVID-19 recovered subjects and 30 individuals vaccinated with both doses of Covishield™. The clinical and demographic details are shown in Table 2. The median age of the COVID-19 recovered patients was 48.5 years, with a range of 23 to 76 years, while the median age of vaccinated group was 45 years, with a range of 18 to 74 years. The distributions of patients based on their age <65 years and >65 years in both groups are shown in Table 2. Out of the participants, 33.33% and 40% females were found in COVID-19 recovered group and vaccinated group, respectively. Based on disease severity, COVID-19 recovered patients were divided into severe and mild groups. Out of the COVID-19 recovered patients 43.33% were severe and required intensive care unit (ICU) support in the hospital. The most prevalent comorbidities among the COVID-19 recovered patients were obesity (26.67%), diabetes (23.33%), and hypertension (6.67%), and some of the patients required intensive care and intubation due to the severity of their condition. Obesity, hypertension, and diabetes were the most common comorbidities observed in the patients who required ICU support.
| Characteristics | COVID-19 Recovered patients (n = 30) | Individuals vaccinated with both doses of Covishield™ (n = 30) |
|---|---|---|
| Age | ||
| Median (IQR) | 48.5 (23-76) | 45(18-74) |
| <65 years (%) | 21(70) | 18 (60) |
| 65+ years (%) | 9(30) | 12 (40) |
| Female (%) | 10(33.33) | 12 (40) |
| Severity | ||
| Severe (%) | 13(43.33) | NA |
| Mild (%) | 17(66.67) | NA |
| Hospitalized, no ICU (%) | 17 (66.67) | NA |
| Hospitalized, required ICU (%) | 13(43.33) | NA |
| Died due to COVID-19 (%) | 0(0) | NA |
| Preexisting conditions | ||
| No underlying disease | 11(36.67) | 24 (80) |
| Hypertension | 2(6.67) | 2 (6.67) |
| Diabetes | 7(23.33) | 1 (3.33) |
| Obesity | 8(26.67) | 2 (6.67) |
| Hypothyroidism | 2(6.67) | 0 |
ICU = intensive care unit; IQR, interquartile range.
Serum levels of total antibodies in COVID-19 recovered patients and vaccinated individuals
The sensitivity and specificity of human SARS-CoV-2 spike (trimer) Ig total ELISA kit (Invitrogen; ThermoFisher Scientific) were tested by the serum sample collected before 2019 as a control. SARS-CoV-2 spike protein-specific total antibodies were measured 3-4 weeks after the recovery from SARS-CoV-2 infections and after receiving the second dose of Covishield™ vaccine. The median (IQR) serum levels of SARS-CoV-2 spike-specific total antibodies were found to be 1275 U/mL (650-2000 U/mL) and 2100 U/mL (1300-3000 U/mL) in COVID-19 recovered patients and vaccinated individuals, respectively. The serum levels of total antibodies were found to be significantly (p<0.001) higher in the vaccinated individuals as compared to COVID-19 recovered patients (Fig. 1).
Immunoreactivity of peptides with anti-sera of vaccinated and COVID-19 recovered patients
The individual peptides (P1, P3, P5, P7, P9, and P11) of wild-type SARS-CoV-2 and peptides (P2, P4, P6, P8, P10, and P12) were coated in the high-binding microtiter plate to assess the immunoreactivity with anti-sera obtained from vaccinated people and COVID-19 recovered patients. The immunoreactivity of peptides with antibodies was observed by taking the optical density (OD) at 592 nm. The difference in the OD value with corresponding peptides of wild types and mutated peptides was reported. The significant (p<0.05) difference in the immunoreactivity of peptides of wild type (P1, P5, P7) with peptides (P2, P6, P8) of mutated SARS-CoV-2 is reported in Figure 2. As compared to wild-type peptide sequence (P1), the mutated peptide sequence (P2) has single amino acid mutation in the RBD at position 417, which significantly reduced the immunoreactivity with anti-sera of vaccinated people and COVID-19 recovered patients (Fig. 2A). Likewise, the peptide sequence (P6) has a single amino acid mutation at residue 484 in the RBD of SARS-CoV-2, which significantly (p<0.05) reduced the immunoreactivity with anti-sera of vaccinated people and COVID-19 recovered patients (Fig. 2B). Similar to the mutated peptide sequences P2 and P6 of SARS-CoV-2 variants, the peptide sequence (P8) has single amino acid mutation in the RBD at the residue 501. This mutation also reduced the immunoreactivity with anti-sera of the vaccinated people and COVID-19 recovered patients (Fig. 2C). Finally, we have tested the immunoreactivity of peptide pools (P1, P5, and P7) of wild-type SARS-CoV-2 and peptide pools (P2, P6, and P8) of SARS-CoV-2 variants with the anti-sera of vaccinated people and COVID-19 recovered patients. Our finding showed a significant (p<0.05) reduction in immunoreactivity of anti-sera of vaccinated and COVID-19 recovered patients with peptide pools of SARC-CoV-2 variants (Fig. 2D). The amino acid mutations in the RBD of SARS-CoV-2 variants at residues 477, 478 for peptide sequence P4 and at residues 400, 446 for peptide sequence P10 did not reduce the immunoreactivity with anti-sera of vaccinated people and COVID-19 recovered patients (Supplementary figure 1, a and b). However, the mutated peptide sequence (P12) of SARS-CoV-2 variants has five amino acid mutations at residues 493, 496, 498, 501, and 505, which reduced the immunoreactivity with anti-sera of vaccinated people and COVID-19 recovered patients. But, it was found to be comparable (p>0.05) (Supplementary figure 1c).
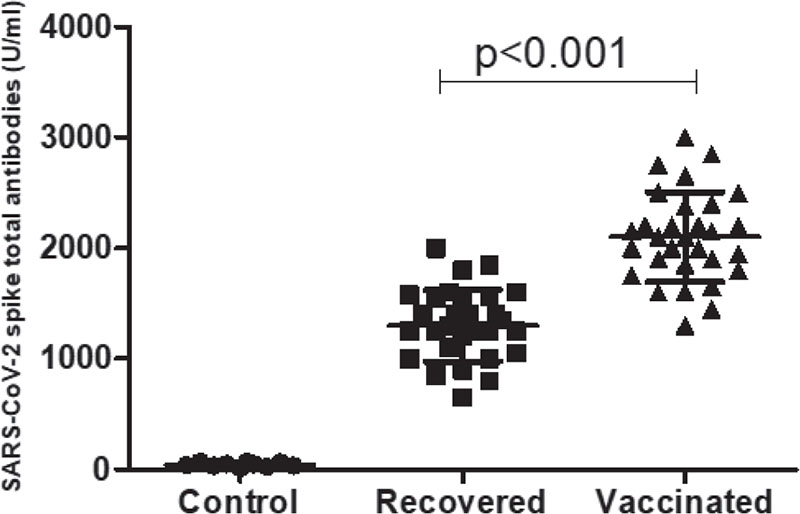
FIGURE 1 - Serum levels of SARS-CoV-2 spike protein-specific total immunoglobulins (Ig) within 3-4 weeks of recovery from SARS-CoV-2 infections from the hospital (n = 30) and 3-4 weeks after receiving the second dose of Covishield™ vaccine.
Finally, our study reported that the mutations in the RBD at the residues K417, E484, and N501 in SARS-CoV-2 variants have been associated with reduced immunoreactivity with anti-sera of vaccinated people and COVID-19 recovered patients (Fig. 2). The amino acid substitutions at K417, E484, and N501 residues significantly reduced antibody binding with polyclonal serum antibodies obtained from people who have recovered from COVID-19 and also vaccinated with ChAdOx1 nCoV-19 vaccines. The findings indicate that vaccination may result in a decreased long-term antibody response to Beta and Omicron RBDs, highlighting the challenges in neutralizing these variants with the existing antibody response induced by vaccination.
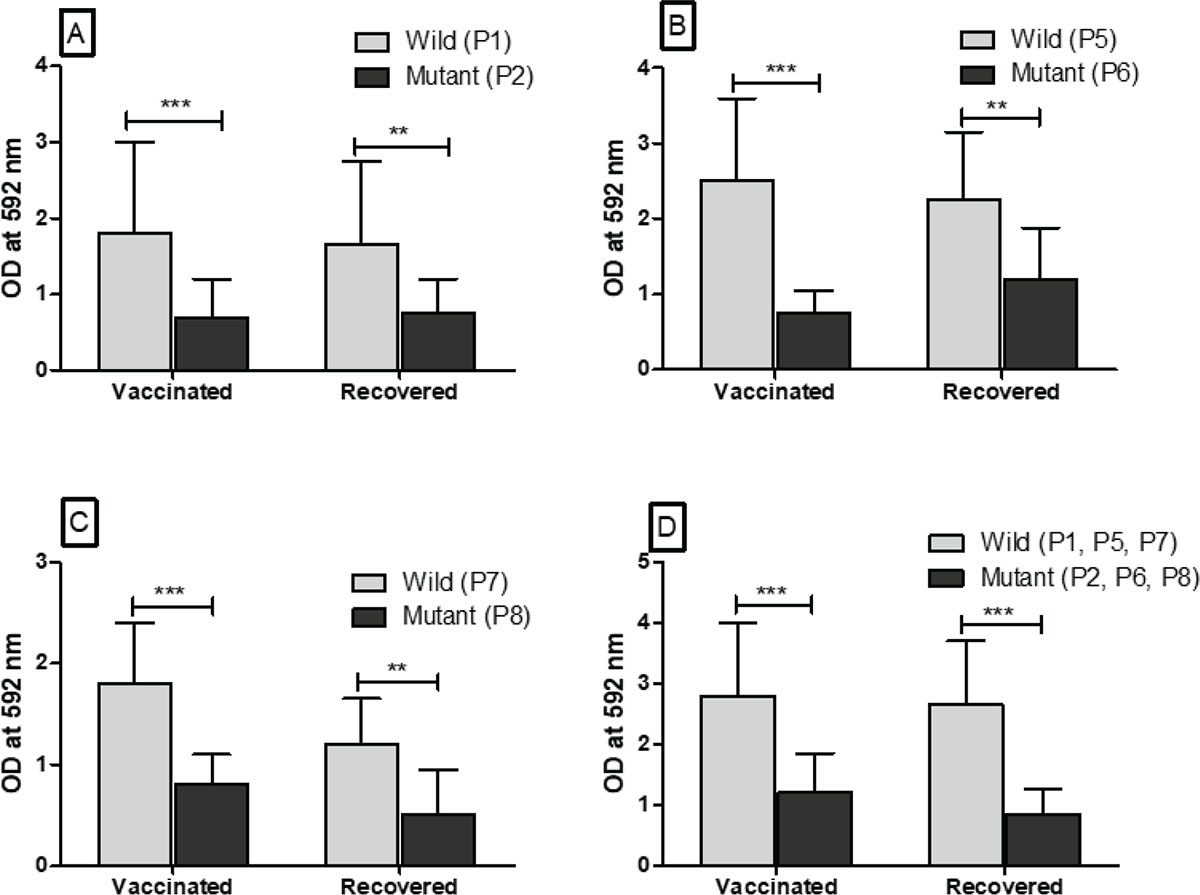
FIGURE 2 - Immunoreactivity of peptides (P1, P5, P7) of wild-type SARS-CoV-2 and peptides (P2, P6, P8) of SARS-CoV-2 variants with anti-sera obtained from healthy individuals vaccinated with both doses of Covishield™ (n = 30) and COVID-19 recovered patients from the hospital (n = 30). A, B, and C. The immunoreactivity of anti-sera with each peptide (P1, P5, P7) of wild-type SARS-CoV-2 and each peptide (P2, P6, P8) of mutated SARS-CoV-2 having single amino acid mutation at residues 417, 484, and 501, respectively. D. The immunoreactivity of anti-sera with peptide pools (P1, P5, P7) of wild-type SARS-CoV-2 and peptide pools (P2, P6, P8) of mutated SARS-CoV-2 having mutations at all three residues 417, 484, and 501.
Discussion
We analyzed the antibody response against SARS-CoV-2 spike protein using blood samples collected at least 3-4 weeks after recovery from nonvaccinated individuals who had recovered from COVID-19. The serum levels of total antibody were compared with individuals who had received both doses of Covishield™. The findings of the present study showed the significant rise in the levels of total antibody after 3-4 weeks of recovery from SARS-CoV-2 infections and after receiving both doses of Covishield™ (ChAdOx1 nCoV-19 coronavirus vaccines). The raised level of total antibody titer was found to be significantly higher in vaccinated people compared to nonvaccinated people who have recovered from SARS-CoV-2 infections. Several studies have reported a significant rise in serum levels of total antibodies postvaccination with Covishield™ in healthy individuals (20,21). Few studies reported a significant difference in antibody titer with disease severity among COVID-19 recovered patients (22,23). Interestingly, among the recovered patients, those who had severe COVID-19 had higher levels of anti-spike total antibody compared to those with mild disease (24). Our study also reported significantly higher levels of SARS-CoV-2 spike protein-specific total antibody in severe COVID-19 recovered patients.
Vaccination with ChAdOx1 nCoV-19 coronavirus vaccines led to a significant increase in anti-spike antibody titers, indicating a robust humoral immune response. However, the raised antibody after vaccination could not immune-react with SARS-CoV-2 variant peptides. The polyclonal antibody response postvaccination showed limitations in binding the mutant peptides of RBDs of the Delta and Omicron variants, suggesting a potential challenge in achieving full protection against these variants (25). Findings of this study suggest that vaccination elicits a lower titer of immune-reactive antibodies against the Delta and Omicron RBDs, indicating potential challenges in neutralizing these variants. Overall, the results highlight the need for ongoing monitoring and adaptation of vaccines to address the evolving SARS-CoV-2 variants and their impact on antibody binding. The findings of our study are supported by the study of Geers et al 2021 (26). The study revealed structural analysis that the Delta and Omicron variants share a common immune escape strategy involving specific residues (K417-E484-N501), allowing them to evade neutralization by anti-RBD antibodies (8,26). The study suggests that through mutations of this triad, SARS-CoV-2 has evolved to evade neutralization by the class I/II anti-RBD antibody fraction of hybrid immunity plasma. The findings indicate that vaccination may result in a decreased long-term antibody response to Delta and Omicron RBDs, highlighting the challenges in neutralizing these variants with the existing antibody response induced by vaccination. The study also found that the interval between vaccination and blood draw did not significantly impact antibody levels, indicating the stability of the antibody response over time. These findings highlight the need for continued monitoring and adaptation of vaccines to address the evolving SARS-CoV-2 variants and their immune evasion strategies.
Findings of our study reported reduced immunoreactivity of anti-sera of vaccinated people and COVID-19 recovered patients with a peptide having E484K mutation in the RBD of SARS-CoV-2 variants. Our finding is supported by the study done in Brazilian/Japanese variants (P.2 and P.1) with the E484K mutation, which had significantly decreased neutralization, suggesting evasion of antibody responses (27). The neutralization of the South African B.1.351 strain, which has three mutations in the RBD and has been found in reinfection cases, was significantly reduced for two-dose vaccines, similar to other distantly related coronaviruses, indicating that a small number of mutations can lead to potent humoral immune response escape (28,29). A considerable percentage of recipients did not have detectable neutralization of the B.1.351 variant after receiving two doses of either vaccine (22). Individuals with prior COVID-19 infection or significant exposure had higher neutralization titers and cross-neutralization against various variants, suggesting a combination of prior infection and vaccination may result in broader neutralizing antibody responses (30). The B.1.351 variant of SARS-CoV-2, also known as the South African variant, exhibits resistance to neutralization primarily because of mutations in the RBD of the spike protein. The RBD mutations in B.1.351, including K417N, E484K, and N501Y, contribute to the observed escape from vaccine-induced neutralization (31,32). Variants like P.2 and P.1 with specific RBD mutations reduce neutralization potency and may explain reinfection cases (33).
Conclusion
The study concludes that multiple variants of SARS-CoV-2, including the Delta and Omicron variants, reduced immunoreactivity with polyclonal sera by vaccine-induced humoral immunity. The Delta variant’s neutralization is primarily attributed to mutations in the RBD of the spike protein. The RBD mutations, such as K417N, E484K, and N501Y, play a significant role in the escape from vaccine-induced neutralization. The study highlights the need for reformulating existing vaccines to include diverse spike sequences and the development of new vaccines capable of eliciting broadly neutralizing antibodies to effectively combat the infection with variants of SARS-COV-2 and possible future pandemic. It also emphasizes the importance of assessing other antibody-mediated functions and cellular immune responses mediated by T cells and natural killer (NK) cells in understanding the overall protection provided by vaccines.
Acknowledgments
We would like to acknowledge the All India Institute of Medical Science, Raipur, for providing the financial support for the study via intramural grant (No.22/41/2019/Admin/1346).
Disclosures
Conflict of interest: All the authors of the manuscript do not have any conflict.
Financial support: The study was supported by intramural research grant sanctioned by All India Institute of Medical Sciences, Raipur, Chhattisgarh, India.
References
- 1. To KKW, Sridhar S, Chiu KHY, et al. Lessons learned 1 year after SARS-CoV-2 emergence leading to COVID-19 pandemic. Emerg Microbes Infect. 2021;10(1):507-535. CrossRef PubMed
- 2. Khan M, Adil SF, Alkhathlan HZ, et al. COVID-19: a global challenge with old history, epidemiology and progress so far. Molecules. 2020;26(1):39. CrossRef PubMed
- 3. Tregoning JS, Flight KE, Higham SL, Wang Z, Pierce BF. Progress of the COVID-19 vaccine effort: viruses, vaccines and variants versus efficacy, effectiveness and escape. Nat Rev Immunol. 2021;21(10):626-636. CrossRef PubMed
- 4. Tsurkalenko O, Bulaev D, O’Sullivan MP, et al; CON-VINCE consortium and the ORCHESTRA working group. Creation of a pandemic memory by tracing COVID-19 infections and immunity in Luxembourg (CON-VINCE). BMC Infect Dis. 2024;24(1):179. CrossRef PubMed
- 5. Akhlaghi A, Darabi A, Mahmoodi M, et al. The frequency and clinical assessment of COVID-19 in patients with chronic rhinosinusitis. Ear Nose Throat J. 2024;103(2):NP98-NP103. CrossRef PubMed
- 6. Su S, Zhao Y, Zeng N, et al. Epidemiology, clinical presentation, pathophysiology, and management of long COVID: an update. Mol Psychiatry. 2023;28(10):4056-4069. CrossRef PubMed
- 7. Zaidi AK, Singh RB. Epidemiology of COVID-19. Prog Mol Biol Transl Sci. 2024;202:25-38. PubMed
- 8. Montezano AC, Camargo LL, Mary S, et al. SARS-CoV-2 spike protein induces endothelial inflammation via ACE2 independently of viral replication. Sci Rep. 2023;13(1):14086. CrossRef PubMed
- 9. Kibria MG, Lavine CL, Tang W, et al. Antibody-mediated SARS-CoV-2 entry in cultured cells. EMBO Rep. 2023;24(12):e57724. CrossRef PubMed
- 10. Li CJ, Chang SC. SARS-CoV-2 spike S2-specific neutralizing antibodies. Emerg Microbes Infect. 2023;12(2):2220582. CrossRef PubMed
- 11. Akaishi T, Fujiwara K. Insertion and deletion mutations preserved in SARS-CoV-2 variants. Arch Microbiol. 2023;205(4):154. CrossRef PubMed
- 12. Ma KC, Shirk P, Lambrou AS, et al. Genomic surveillance for SARS-CoV-2 variants: circulation of omicron lineages – United States, January 2022-May 2023. MMWR Morb Mortal Wkly Rep. 2023;72(24):651-656. CrossRef PubMed
- 13. Aleem A, Akbar Samad AB, Vaqar S. Emerging variants of SARS-CoV-2 and novel therapeutics against coronavirus (COVID-19). In: StatPearls. [Internet] StatPearls Publishing; 2024, Available from Online. Accessed March 1, 2024.
- 14. Roemer C, Sheward DJ, Hisner R, et al. SARS-CoV-2 evolution in the Omicron era. Nat Microbiol. 2023;8(11):1952-1959. CrossRef PubMed
- 15. Carabelli AM, Peacock TP, Thorne LG, et al; COVID-19 Genomics UK Consortium. SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nat Rev Microbiol. 2023;21(3):162-177. CrossRef PubMed
- 16. Pather S, Madhi SA, Cowling BJ, et al. SARS-CoV-2 Omicron variants: burden of disease, impact on vaccine effectiveness and need for variant-adapted vaccines. Front Immunol. 2023;14:1130539. CrossRef PubMed
- 17. Yadav T, Kumar S, Mishra G, Saxena SK. Tracking the COVID-19 vaccines: the global landscape. Hum Vaccin Immunother. 2023;19(1):2191577. CrossRef PubMed
- 18. Wahid M, Jawed A, Mandal RK, et al. Role of available COVID-19 vaccines in reducing deaths and perspective for next generation vaccines and therapies to counter emerging viral variants: an update. Minerva Med. 2023;114(5):683-697. CrossRef PubMed
- 19. Bostanghadiri N, Ziaeefar P, Mofrad MG, Yousefzadeh P, Hashemi A, Darban-Sarokhalil D. COVID-19: An overview of SARS-CoV-2 variants – the current vaccines and drug development. BioMed Res Int. 2023;2023:1879554. CrossRef PubMed
- 20. Nanda R, Gupta P, Giri AK, Patel S, Shah S, Mohapatra E. Serological evaluation of antibody titers after vaccination against COVID-19 in 18-44-year-old individuals at a tertiary care center. Cureus. 2023;15(6):e40543. CrossRef PubMed
- 21. Dimitroff SJ, Würfel L, Meier M, et al. Estimation of antibody levels after COVID-19 vaccinations: preliminary evidence for immune interoception. Biol Psychol. 2023;182:108636. CrossRef PubMed
- 22. Huang C, Huang L, Wang Y, et al. 6-Month consequences of COVID-19 in patients discharged from hospital: a cohort study. Lancet. 2023;401(10393):e21-e33. CrossRef PubMed
- 23. Amrutha S, Kaur K, Aggarwal D, Sodhi MK, Jaswal S, Saini V. Serial evaluation of antibody titres in patients recovered from COVID-19 and their correlation with disease severity. Monaldi Arch Chest Dis 2023 Nov 3. CrossRef
- 24. de Oliveira MI, Aciole MR, Neves PAF, et al. A stronger antibody response in increased disease severity of SARS-CoV-2. BMC Infect Dis. 2024;24(1):17. CrossRef PubMed
- 25. Lavezzo E, Pacenti M, Manuto L, et al. Neutralising reactivity against SARS-CoV-2 delta and omicron variants by vaccination and infection history. Genome Med. 2022;14(1):61. CrossRef PubMed
- 26. Geers D, Shamier MC, Bogers S, et al. SARS-CoV-2 variants of concern partially escape humoral but not T-cell responses in COVID-19 convalescent donors and vaccinees. Sci Immunol. 2021 May 25;6(59):eabj1750. CrossRef
- 27. Ma QL, Huang FM, Guo W, Feng KY, Huang T, Cai YD. Machine learning classification of time since BNT162b2 COVID-19 vaccination based on array-measured antibody activity. Life (Basel). 2023;13(6):1304. CrossRef PubMed
- 28. Edara VV, Norwood C, Floyd K, et al. Reduced binding and neutralization of infection- and vaccine-induced antibodies to the B.1.351 (South African) SARS-CoV-2 variant. bioRxiv [Preprint]. 2021 Feb 22. CrossRef PubMed
- 29. Ryu DK, Song R, Kim M, et al. Therapeutic effect of CT-P59 against SARS-CoV-2 South African variant. Biochem Biophys Res Commun. 2021;566:135-140. CrossRef PubMed
- 30. Sette A, Crotty S. Immunological memory to SARS-CoV-2 infection and COVID-19 vaccines. Immunol Rev. 2022;310(1):27-46. CrossRef PubMed
- 31. Viana R, Moyo S, Amoako DG, et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 2022 Mar;603(7902):679-686. CrossRef
- 32. Wibmer CK, Ayres F, Hermanus T, et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med. 2021 Mar 1;27:622-625.
- 33. Wahid M, Jawed A, Mandal RK, et al. Variants of SARS-CoV-2, their effects on infection, transmission and neutralization by vaccine-induced antibodies. Eur Rev Med Pharmacol Sci. 2021;25(18):5857-5864. PubMed