 |
Drug Target Insights 2023; 17: 101-109 ISSN 1177-3928 | DOI: 10.33393/dti.2023.2638 ORIGINAL RESEARCH ARTICLE |
 |
Identification of anti-pathogenic activity among in silico predicted small-molecule inhibitors of Pseudomonas aeruginosa LasR or nitric oxide reductase (NOR)
ABSTRACT
Introduction: Antibiotic-resistant Pseudomonas aeruginosa strains cause considerable morbidity and mortality globally. Identification of novel targets in this notorious pathogen is urgently warranted to facilitate discovery of new anti-pathogenic agents against it. This study attempted to identify small-molecule inhibitors of two important proteins LasR and nitric oxide reductase (NOR) in P. aeruginosa. ‘Las’ system can be said to be the ‘master’ regulator of quorum sensing in P. aeruginosa, whose receptor protein is LasR. Similarly, NOR is crucial to detoxification of reactive nitrogen species.
Methods: In silico identification of potential LasR or NOR inhibitors was attempted through a virtual screening platform AtomNet® to obtain a final subset of <100 top scoring compounds. These compounds were evaluated for their in vivo anti-pathogenic activity by challenging the model host Caenorhabditis elegans with P. aeruginosa in the presence or absence of test compounds. Survival of the worm population in 24-well assay plates was monitored over a period of 5 days microscopically.
Results: Of the 96 predicted LasR inhibitors, 11 exhibited anti-Pseudomonas activity (23%-96% inhibition of bacterial virulence as per third-day end-point) at 25-50 µg/mL. Of the 85 predicted NOR inhibitors, 8 exhibited anti-Pseudomonas activity (40%-85% inhibition of bacterial virulence as per second-day end-point) at 25-50 µg/mL.
Conclusion: Further investigation on molecular mode of action of compounds found active in this study is warranted. Virtual screening can be said to be a useful tool in narrowing down the list of compounds requiring actual wet-lab screening, saving considerable time and efforts for drug discovery.
Keywords: Antimicrobial resistance (AMR), Nitric oxide, Nitrosative stress, Priority pathogen, Pseudomonas aeruginosa, Quorum sensing (QS), Virulence
Received: July 20, 2023
Accepted: September 4, 2023
Published online: September 28, 2023
This article includes supplementary material
Corresponding author:
Vijay Kothari
Institute of Science, Nirma University
S-G Highway, Ahmedabad-382481 - India
vijay23112004@yahoo.co.in; vijay.kothari@nirmauni.ac.in
Bryan Cox’s affiliation at the time of study was Atomwise Inc.
Drug Target Insights - ISSN 1177-3928 - www.aboutscience.eu/dti
© 2023 The Authors. This article is published by AboutScience and licensed under Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0).
Commercial use is not permitted and is subject to Publisher’s permissions. Full information is available at www.aboutscience.eu
Introduction
Antimicrobial resistance (AMR) among infectious bacteria has emerged as a healthcare challenge of global concern (Online). The World Health Organization (WHO) has also published a global action plan on AMR in 2015. As per the Indian National Action Plan on Antimicrobial Resistance (NAP-AMR: 2017-2021), India is among the nations with the highest burden of bacterial infections. The crude mortality from infectious diseases in India today is 417 per 100,000 persons. The situation in other developing countries is equally grave. Murray et al estimated 1.27 million deaths attributable to bacterial AMR in 2019 (1). Of the six leading pathogens identified by them responsible for maximum death toll, one is Pseudomonas aeruginosa. It is among the most notorious pathogenic bacteria, and its carbapenem-resistant phenotype has been listed by the WHO among priority pathogens for the development of new antibiotics (Online). Antibiotic-resistant P. aeruginosa has been listed as an important pathogen by the U.S. Centers for Disease Control and Prevention (CDC) (Online) as well as the Department of Biotechnology, India (DBT) (Online) against which new antimicrobials are urgently required.
The pipeline for new antibiotics does not contain sufficient number of promising candidates (2). There is a dearth of novel antibacterial leads as well as targets (3,4). Identification and validation of new targets in important pathogens is of utmost significance (5,6). Conventional bactericidal antibiotics attack a very narrow range of targets in susceptible bacteria, for example, cell wall synthesis, protein synthesis, nucleic acid synthesis, etc. Since the past decade, there has been much interest in the research community regarding discovery of anti-virulence molecules, which may attenuate the bacterial virulence without necessarily killing them. Such ‘pathoblockers’ are expected to compromise the ability of the pathogen to damage the host, by exerting their effect on nonessential targets such as bacterial quorum sensing (QS) (7), stress-response machinery (8), metal homeostasis (9,10), etc. The next-generation antibacterials preferably should be attacking such hitherto unexplored/underexplored targets in pathogenic bacteria (11-13).
Proper functioning of the chemical signal-based intercellular communication, known as QS, is crucial for sufficient expression of virulence in pathogens like P. aeruginosa, as QS is an effective mechanism for regulating expression of multiple genes associated with a multitude of functions (14). Interrupting bacterial QS can be an effective strategy to combat pathogens (15,16). QS consists of two components: signal generation and signal response, respectively, encoded by LuxI and LuxR homologues (17,18). Inhibiting the ‘signal response’ component of QS (e.g. LasR in P. aeruginosa) can notably compromise their ability to exert collective behaviour in response to environmental changes or host defence. P. aeruginosa regulates its drug resistance and pathogenicity through multiple QS mechanisms including the LasI/R, RhlI/R, and PQS/MvfR systems. Targeting one or more of these QS systems may prove an effective way of dealing with P. aeruginosa infections (19). Owing to the important role of Las system in overall QS circuit of P. aeruginosa, its receptor protein LasR is believed to be a plausible anti-virulence target (20). LasR is a transcriptional activator of various virulence-associated genes in P. aeruginosa, which recognizes a specific signal molecule, namely N-(3-oxo-dodecanoyl)-L-homoserine lactone (3O-C12-HSL) (21). The LasR-3O-C12-HSL complex triggers the expression of multiple QS-regulated genes. Potential LasR inhibitors either may prevent its binding with its natural ligand or compromise its ability to affect expression of target genes (22). QS not being essential for bacterial survival, its inhibitors are expected to exert lesser selection pressure on bacterial population with respect to development of resistant phenotypes (23). Additionally, due to the overlap in QS systems among various gram-negative bacteria (24-25), inhibitors effective against one gram-negative species may have broad-spectrum activity against multiple other gram-negative pathogens.
Pathogens striving to survive inside a host body are forced to face a variety of stresses such as iron deprivation, oxidative stress, nitrosative stress, etc. Bacteria employ antioxidant enzymes to counter reactive oxygen species, and similarly certain other enzymes to counter reactive nitrogen species (26). From the work done in our lab as well as literature survey, we consider the components of P. aeruginosa genome involved in responding to nitrosative stress to be potential targets. Among the components of nitrosative stress response in P. aeruginosa, one important enzyme is nitric oxide reductase (NOR). This protein was one of the major targets of an anti-infective polyherbal formulation (Panchvalkal) investigated by us in the recent past (6,27). NOR also emerged among the top differentially expressed genes in P. aeruginosa treated by us with other anti-virulence polyherbal formulations Enteropan (SRX15248092) or colloidal silver (SRX14392191) at sub-lethal levels.
NOR is an important detoxifying enzyme in P. aeruginosa, which is crucial to its ability to withstand nitrosative stress (e.g. in the form of nitric oxide [NO]). NOR has also been reported (28,29) to be important for virulence expression of this pathogen, and thus can be a plausible target for novel anti-virulence agents. Molecules capable of inhibiting NOR can be expected to compromise the pathogen’s ability to detoxify NO, not allowing its virulence traits (e.g. biofilm formation, as NO has been indicated to act as a biofilm-dispersal signal) to be expressed fully (30). The test molecules capable of inhibiting NOR may emerge as novel anti-biofilm agents not only against P. aeruginosa but against multiple pathogens as NO is reported to be perceived as a dispersal signal by various gram-negative and gram-positive bacteria (31). Thus, NOR inhibitors may be expected to have a broad-spectrum activity against multiple pathogens. Major function of NOR is to detoxify NO generated by nitrite reductase (NIR). NO is a toxic by-product of anaerobic respiration in P. aeruginosa. NO-derived nitrosative species can damage deoxyribonucleic acid (DNA) and compromise protein function. Intracellular accumulation of NO is likely to be lethal for the pathogen (32). It can be logically anticipated that P. aeruginosa’s ability to detoxify NO will be compromised under the influence of a potent NOR inhibitor. Since NO seems to have a broad-spectrum anti-biofilm effect, NOR activity is essential for effective biofilm formation by the pathogens. NOR activity and NO concentration can modulate cellular levels of cyclic di-GMP, which is a secondary messenger molecule recognized as a key bacterial regulator of multiple processes such as virulence, differentiation, and biofilm formation (33). In mammalian pathogens, the host’s macrophages are a likely source of NO. NOR expressed by the pathogen provides protection against the host defence mechanism. Since NOR activity is known to be important in multiple pathogenic bacteria (e.g. P. aeruginosa, Staphylococcus aureus, Serratia marcescens) for biofilm formation, virulence expression, combating nitrosative stress, and evading hose defence, NOR seems to be an important target for novel anti-pathogenic agents. Any molecule capable of interfering with bacterial NOR activity is likely to be an effective anti-pathogenic agent, since bacterial populations require NOR for various purposes including detoxification and evasion of host defences (34). A potential NOR inhibitor besides troubling the pathogen directly may also boost its clearance by the host macrophages.
Proteins such as LasR and NOR important to the pathogens and whose structure is known can be useful starting point for a drug discovery programme. In silico virtual screening tools can be used to screen large chemical libraries to predict inhibitors of the target proteins. In the recent past, quite a few potent anti-pathogenic compounds have been identified using this approach. For example, one such virtual screening study by Abelyan et al (35) identified benzamides, synthetic derivatives of flavones, as potential inhibitors of LasR. Another in silico effort by Narayanaswamy et al (36) identified potent inhibitors of enzymes involved in nitrogen metabolism in various bacteria including P. aeruginosa, nitrous oxide reductase (N2OR), and NIR, from among a library of synthetic and natural compounds.
This study aimed at screening 96 compounds identified through a virtual screen as potential LasR inhibitors, and 85 compounds predicted to be NOR inhibitors in silico for their possible anti-virulence activity against P. aeruginosa in the model host Caenorhabditis elegans. Any such potent NOR or LasR inhibitors identified through this study may prove to be useful lead(s) for novel anti-pathogenic drug development. They can be useful either as standalone therapy or in combination with conventional antibiotics.
Methods
Virtual screening
The virtual screening from a library of approximately 3 million compounds was conducted using AtomNet® screening platform (37). AtomNet® is a proprietary deep learning neural network useful for structure-based drug design and discovery through its small-molecule binding affinity prediction capacity.
Screening for LasR binding ligands
There are a number of available crystal structures of LasR in complex with small molecules, including the endogenous ligand and other agonists. Although discovery of a potent antagonist is preferable, the virtual screen attempted to find novel chemical matter that binds at the desired site and the mode of binding may be analysed later. All of the available crystal structures were considered as receptor templates for virtual screening. The highest resolution structure, PDB 3IX3, chosen as the ligand binding pocket is deep and solvent excluded and appears well-suited for binding small molecules (Fig. 1). The screening volume was restricted to the binding pocket surrounding the existing ligand. A screening library of approximately 3 million compounds was exhaustively pose-sampled and scored, followed by filtering for drug-like properties, and selection of the top 96 compounds for ordering in physical form.
Screening for NOR binding ligands
The structure of P. aeruginosa NOR bound to an antibody fragment has been determined by crystallography (PDBID 3O0R). This structure reveals NOR composed of two subunits: NORB containing 12 transmembrane helices and NORC with a transmembrane helix and hydrophilic periplasmic domain. Three haem cofactors are complexed in NOR. Two haems are deep in the NORB transmembrane region, and the third haem spans the NORB-NORC interface. No drug-like small-molecule inhibitors have been reported for NOR, and no druggable pocket is obvious from the reported structure. To identify potential targetable sites, the structure of NOR was analysed by fPocket. This analysis revealed a pocket at the interface of NORB and NORC and near one of the NORB haem cofactors. This positioning suggests that small-molecule binding to this pocket may disrupt norB-norC interactions and/or inhibit haem-mediated electrochemistry. The virtual screen therefore sought to identify small molecules that bind to this pocket in NOR and potentially disrupt enzymatic function. A library of approximately 3 million compounds was screened for identifying compounds capable of virtual binding at the selected target site on NOR (Fig. 2) using AtomNet®. Top scoring compounds were clustered and filtered to arrive at a final subset of 85 deliverable compounds.
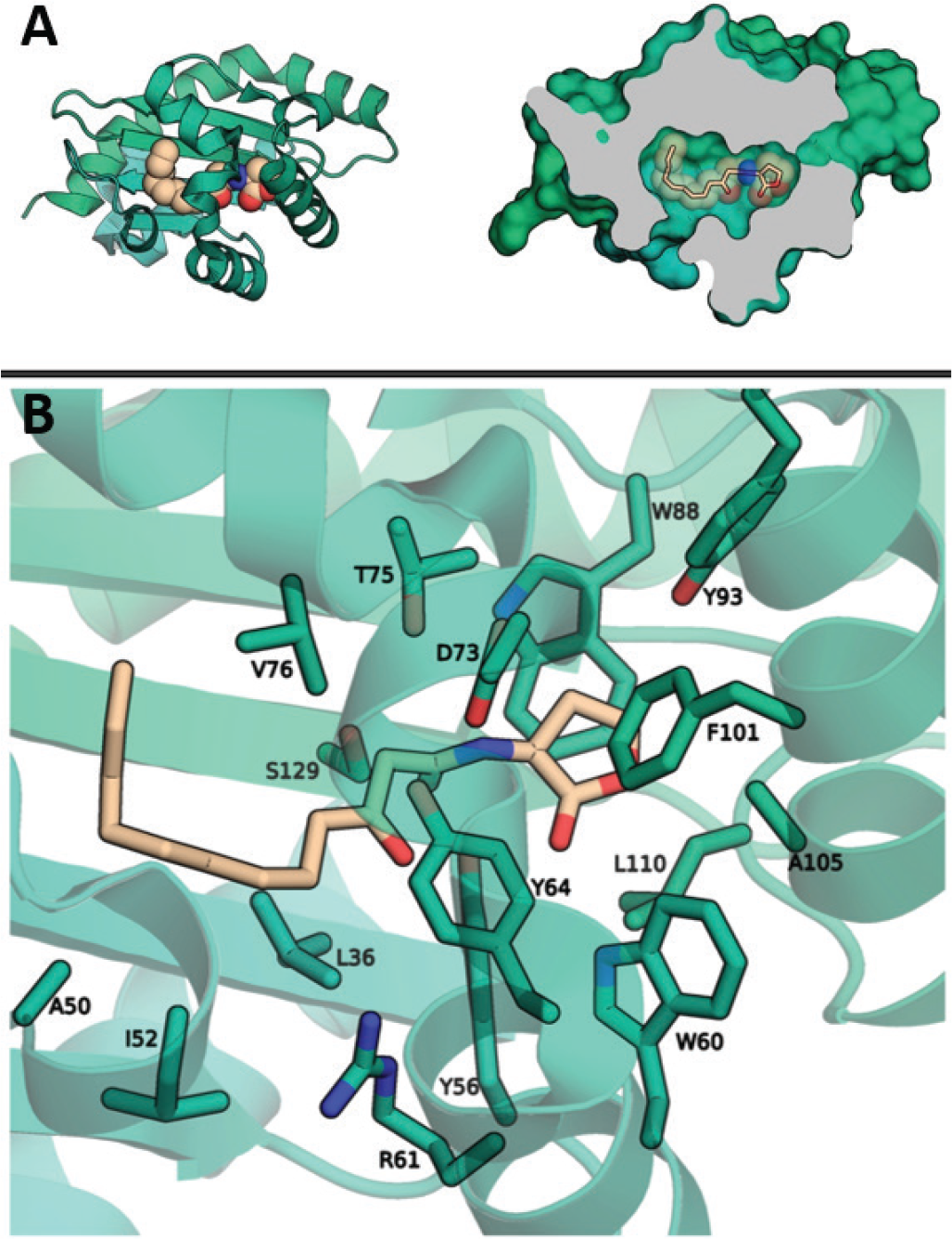
Fig. 1 - Quorum sensing receptor LasR. A) The structure of LasR (green) in complex with its natural ligand 3O-C12-HSL (tan). Coordinates taken from PDB 3IX3. B) The binding site of LasR with the 3O-C12-HSL ligand is present. Residues expected to make direct contacts with ligands are labelled.
Test compounds
Ninety-six compounds showing in silico affinity to LasR (Supplementary table S1, List of predicted LasR inhibitors subjected to wet-lab assay) and 85 compounds showing in silico affinity to NOR (Supplementary table S2, List of predicted NOR inhibitors subjected to wet-lab assay) were ordered in physical form from Enamine (Kyiv, Ukraine) and Mcule (Budapest, Hungary), respectively. Test compounds were stored in the refrigerator and reconstituted in dimethyl sulfoxide (DMSO; 500-1000 µL) (Merck) on the day of assay.
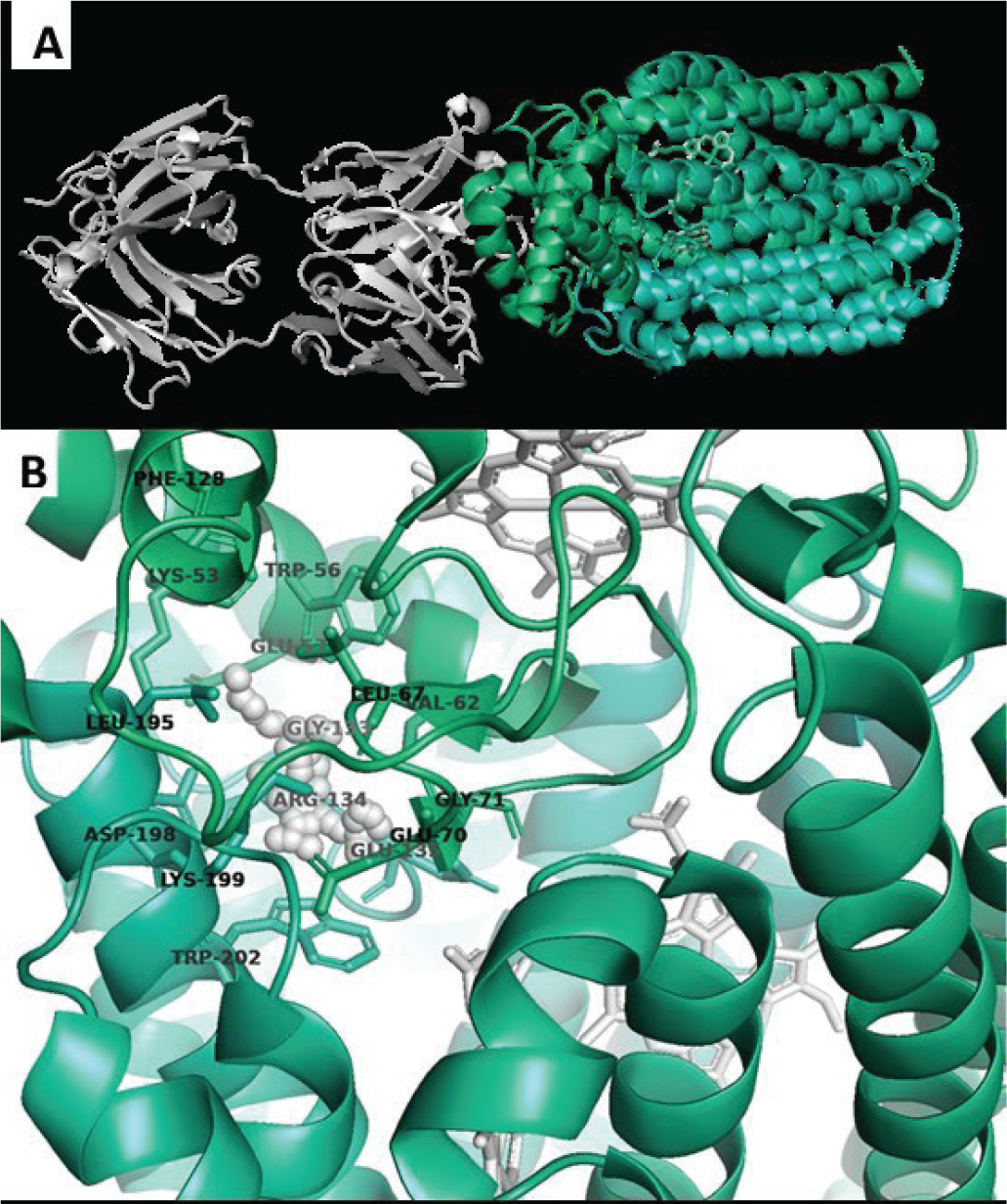
Fig. 2 - Nitric oxide reductase (NOR). A) Crystal structure of NOR complexed with an antibody fragment (white cartoons) and with three haem cofactors (white sticks). B) Proposed target for virtual screen (white spheres) with surrounding residues from NOR chains B and C showing the proximity to the haem cofactors.
Bacterial strain
The P. aeruginosa strain used in this study was sourced from our internal culture collection, which has been characterized by us for its antibiotic susceptibility/resistance, pigment production, and certain other virulence traits. Its antibiogram (Online) generated through a disc-diffusion assay performed as per the National Committee for Clinical Laboratory Standards (NCCLS) guidelines revealed it to be resistant to eight antibiotics (cotrimoxazole, augmentin, nitrofurantoin, ampicillin, chloramphenicol, clindamycin, cefixime, and vancomycin) belonging to five different classes. Hence it can be described as a multidrug-resistant (MDR) strain. As reported in our earlier publications (27,38) with this strain, it is a haemolytic strain capable of producing the QS-regulated pigments (pyocyanin and pyoverdine), and also of biofilm formation. We maintained this bacterium on Pseudomonas agar (HiMedia). While culturing the bacteria for in vivo assay, they were grown in Pseudomonas broth (magnesium chloride 1.4 g/L, potassium sulphate 10 g/L, peptic digest of animal tissue 20 g/L, pH 7.0 ± 0.2).
Nematode host
C. elegans (N2 Bristol) was used as the model host in this study. Worms were maintained on nematode growth medium (NGM): 3 g/L NaCl (HiMedia, MB023-500G), 2.5 g/L peptone (HiMedia), 17 g/L agar-agar (HiMedia), 1 M CaCl2 (HiMedia), 1 M MgSO4 (Merck), 5 mg/mL cholesterol (HiMedia), 1 M phosphate buffer of pH 6, agar plate with Escherichia coli OP50 (LabTIE B.V., the Netherlands) as food. For synchronization of the worm population, adult worms from a 4- to 5-day-old NGM plate were first washed with distilled water, and then treated with 1 mL of bleaching solution (water + 4% NaOCl [Merck] +1 N NaOH [HiMedia] in 3:1:1 proportion), followed by centrifugation (1,500 rpm at 22°C) for 1 min. Eggs in the resultant pellet were washed multiple times with sterile distilled water, followed by transfer onto a new NGM plate seeded with E. coli OP50. L3-L4 stage worms appearing on this plate after 2-3 days of incubation at 22°C were used for further experimentation.
Virulence assay
P. aeruginosa grown in Pseudomonas broth (at 35±1°C for 21 hours with intermittent shaking) was allowed to attack C. elegans (L3-L4 stage) in a 24-well plate (HiMedia) in the presence or absence of test compounds, and their capacity to kill the worm population was monitored over a period of 5 days. In each well, there were 10 worms in M9 buffer (3 g/L KH2PO4, 6 g/L Na2HPO4, 5 g/L NaCl), which were challenged with P. aeruginosa by adding 100 µL (OD764 = 1.30) of bacterial culture grown in Pseudomonas broth. Appropriate controls, that is, worms, exposed neither to test compound nor to bacteria; worms exposed to test compound, but not to bacterial pathogens (toxicity control); worms challenged with bacteria in the presence of 0.5% v/v DMSO (vehicle control); and worms challenged with bacteria in the presence of 0.5 µg/mL ofloxacin (positive control) were also included in the experiment. Incubation was carried out at 22°C. The number of dead vs. live worms was counted every day for 5 days by putting the plate with lid under a light microscope 4× objective. Straight non-moving worms were considered to be dead. Plates were gently tapped to confirm lack of movement in the apparently dead worms. On the last day of the experiment, when plates could be opened, their death was also confirmed by prodding them with a straight wire, wherein no movement was taken as confirmation of death.
Statistics
Results reported are means of three replicates. Statistical significance was assessed through a t-test performed using Microsoft Excel. Values of p≤0.05 were considered to be significant.
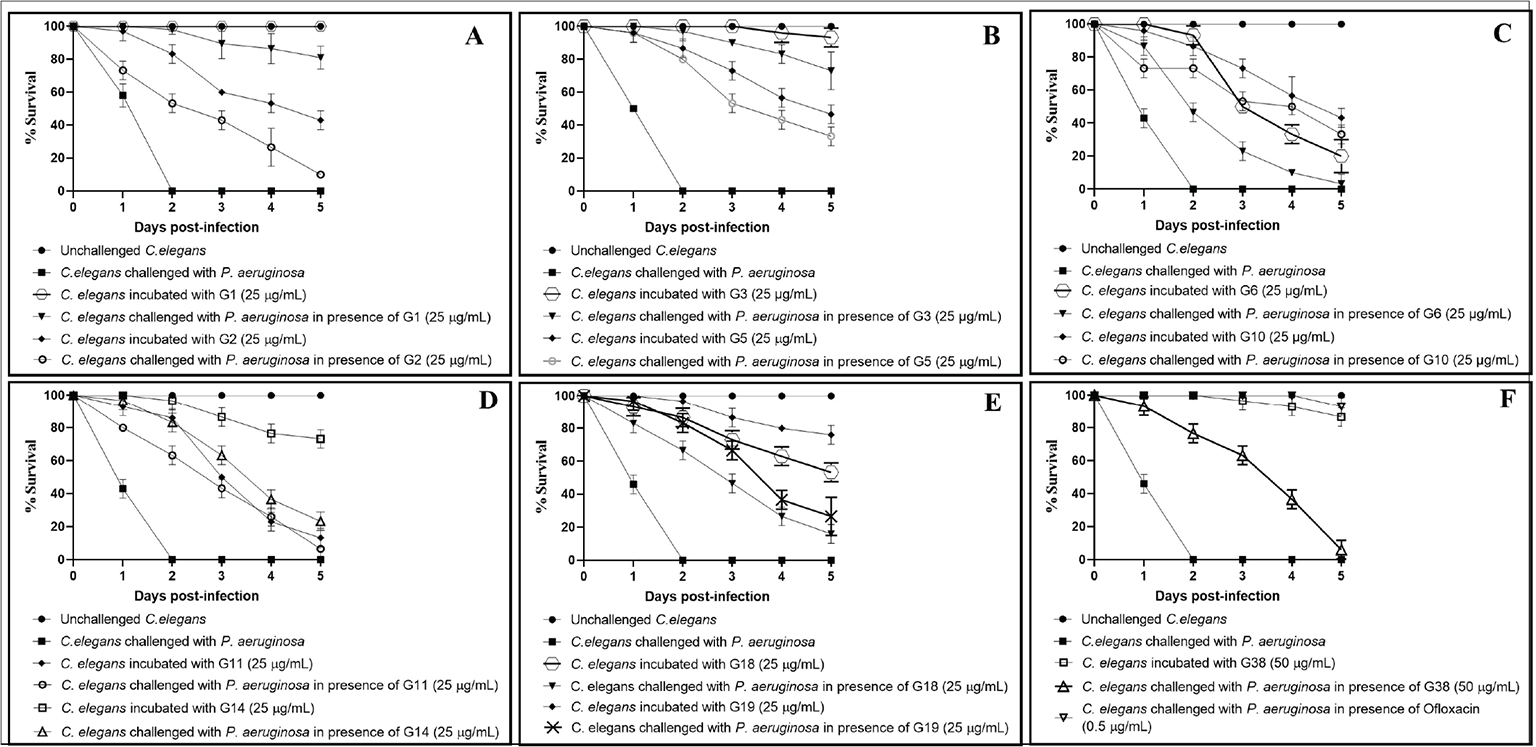
Fig. 3 - Pseudomonas aeruginosa’s virulence towards the host worm gets attenuated in the presence of certain predicted LasR inhibitors. A) P. aeruginosa could kill 90%±9.19%** and 43%±5.7%*** lesser worms in the presence of G1 (Z30981775) and G2 (Z65195564), respectively. B) P. aeruginosa could kill 90%±0%*** and 53%±5.7%*** lesser worms in the presence of G3 (Z212728858) and G5 (Z354444420), respectively. C) P. aeruginosa could kill 23%±5.7%*** and 53%±5.7%*** lesser worms in the presence of G6 (Z400859658) and G10 (Z1426094174), respectively. D) P. aeruginosa could kill 43%±5.7%*** and 63%±5.7%*** lesser worms in the presence of G11 (Z1625994950) and G14 (Z104586200), respectively. E) P. aeruginosa could kill 46%±5.7%*** and 67%±5.7%*** lesser worms in the presence of G18 (Z1084397894) and G19 (Z1212781307), respectively. F) P. aeruginosa could kill 63.3%±5.7%*** and 100%±0%*** lesser worms in the presence of G38 (Z89293640) and ofloxacin (0.5 µg/mL), respectively. Later it was employed as a positive control at its sub-minimum inhibitory concentration (MIC) level, and it did allow progeny formation in worm population from the third day onwards.
Dimethyl sulfoxide (DMSO; 0.5% v/v) present in the ‘vehicle control’ neither affected virulence of the bacterium towards Caenorhabditis elegans, nor did it show any effect on worm survival. **p<0.01, ***p<0.001. The percent values reported pertain to the worm survival 3 days post-infection.
Results
Anti-pathogenic activity of potential LasR inhibitors
Results of anti-virulence assay for all active compounds are presented in Fig. 3. Since P. aeruginosa strain used by us could kill all worms within 18-36 hours, any end-point beyond that can be taken as valid for labelling any compound as ‘active’ or ‘inactive’. However, to have more robust interpretation, we continued worm counting in assay plates till 5 days for comparing number of live worms in experimental vs. control wells.
Eleven of the 88 DMSO-soluble compounds (i.e. 12.5%) assayed exhibited anti-Pseudomonas activity (23%-96% as per third-day end-point) at 25-50 µg/mL (Tab. I). These 11 compounds (G1, G2, G3, G5, G6, G10, G11, G14, G18, G19, and G38) should be tested at still lesser concentrations to find out minimum effective concentration (MEC). Eight of the test compounds were found to possess dual activity, that is, anti-pathogenic as well as anthelmintic. Eight of the active anti-Pseudomonas compounds (G2, G5, G6, G10, G11, G14, G18, G19) identified in this study were also toxic to the host worm at concentrations employed. Hence, they should be tested at still lower concentrations. It is possible that their lower concentrations may exhibit anti-pathogenic activity without exerting any toxicity towards the eukaryotic host. Masking of the anti-pathogenic activity by anti-worm activity of the same compound (Tab. II) needs to be paid attention while interpreting the results. Compounds found to possess anti-pathogenic activity in our study were effective at 25-50 ppm, which seems to be good enough to warrant further investigation, while comparing with effective concentrations reported for other LasR inhibitors. For example, a potent LasR inhibitor, LasR-IN-4, was shown to possess inhibitory activity against P. aeruginosa with MIC of 56.25 μg/mL (39). Another LasR inhibitor, naringenin, was reported to inhibit QS response in P. aeruginosa by competing with N-(3-Oxo-dodecanoyl)-L-homoserine lactone for LasR binding at 136 µg/mL (40). O’Brien et al (41) reported Br-HSL to antagonize LasR with IC50 of 5 μg/mL.
| Lab code | Manufacturer’s code | Conc.(µg/mL) | % reduction in bacterial virulence | |
|---|---|---|---|---|
| 3rd day end-point | 5th day end-point | |||
| G1 | Z30981775 | 25 | 90 ± 9.19** | 82 ± 7.07*** |
| G2 | Z65195564 | 25 | 43 ± 5.7*** | 10 ± 0*** |
| G3 | Z212728858 | 25 | 90 ± 0*** | 73 ± 11.5*** |
| G5 | Z354444420 | 25 | 53 ± 5.7*** | 33 ± 5.7*** |
| G6 | Z400859658 | 25 | 23 ± 5.7*** | 3 ± 5.7 |
| G10 | Z1426094174 | 25 | 53 ± 5.7*** | 33 ± 5.7*** |
| G11 | Z1625994950 | 25 | 43 ± 5.7*** | 6 ± 11.5 |
| G14 | Z104586200 | 25 | 63 ± 5.7*** | 23 ± 5.7*** |
| G18 | Z1084397894 | 25 | 46 ± 5.7*** | 16 ± 5.7** |
| G19 | Z1212781307 | 25 | 67 ± 5.7*** | 26 ± 11.5** |
| G38 | Z89293640 | 50 | 63.3 ± 5.7*** | 6 ± 5.7 |
**p<0.01, ***p<0.001.
| Lab code | Manufacturer’s code | Conc (µg/mL) | % anti-pathogenic activity based on fifth day end-point | |
|---|---|---|---|---|
| Without nullifying compound’s toxicity towards worms | After nullifying compound’s toxicity towards worms | |||
| G2 | Z65195564 | 25 | 10 ± 0*** | 67 ± 0*** |
| G5 | Z354444420 | 25 | 33 ± 5*** | 87 ± 5*** |
| G6 | Z400859658 | 25 | 3 ± 5.7 | 83 ± 5.7*** |
| G10 | Z1426094174 | 25 | 33 ± 5*** | 90 ± 5*** |
| G11 | Z1625994950 | 25 | 6 ± 11.5 | 93 ± 11.5*** |
| G14 | Z104586200 | 25 | 23 ± 5.7** | 50 ± 5.7*** |
| G18 | Z1084397894 | 25 | 16 ± 5.7** | 63 ± 5.7*** |
| G19 | Z1212781307 | 25 | 26 ± 11.5** | 53 ± 11.5*** |
| G38 | Z89293640 | 50 | 6 ± 5.7 | 20 ± 5.7** |
**p<0.01, ***p<0.001.
Further, in vitro incubation of bacteria with the compounds identified in this study to possess anti-P. aeruginosa activity is required to find out whether these compounds exhibit bactericidal/bacteriostatic/anti-virulence activity. While evaluating any compound(s) for their anti-virulence activity, it should be kept in mind that even compounds capable of curbing bacterial virulence partially can be potentially useful in combination with conventional antibiotics. Such compounds may be potential resistance modifiers. Even as a standalone therapy, they may be of indirect help to host immune system by reducing the overall bacterial load to be cleared by the immune system (42,43). Three of these anti-pathogenic compounds (G1, G3, G38) did not exhibit any notable toxicity towards the host worm, and hence seem to be the most logical candidates for further investigation. These compounds should be tested against multiple species of antibiotic-resistant bacteria to know whether they are broad-spectrum antimicrobials. Additionally, their effect on bacterial gene expression at whole transcriptome level should also be investigated to elucidate the underlying molecular mechanisms.
Anti-pathogenic activity of potential NOR inhibitors
Of the total 85 compounds received, 10 were insoluble in the vehicle solvent DMSO. Remaining 75 compounds were assayed for their possible anti-pathogenic activity by challenging the host worm with P. aeruginosa in the presence or absence of test compounds. Eight (~11% of all the compounds tested) of the test compounds were able to rescue the worm population from the pathogen-induced death by 40%-85% (second-day end-point) (Fig. 4; Tab. III).
Five of the test compounds were found to possess dual activity, that is, anti-pathogenic as well as anthelmintic. Five of the active anti-Pseudomonas compounds (N4, N36, N37, N61, N65) identified in this study were also toxic to the host worm at concentrations employed. Hence, they (excluding N61) should be tested at still lower concentrations. It is possible that their lower concentrations may exhibit anti-pathogenic activity without exerting any toxicity towards the eukaryotic host. Masking of the anti-pathogenic activity by anti-worm activity of the same compound (Tab. IV) needs to be paid attention while interpreting the results.
Conclusion
This study is a preliminary demonstration of the utility of virtual screening approach for discovery of potentially novel anti-pathogenic agents. Virtual screening can reduce the number of compounds required to be actually subjected to wet-lab assays, and thus reducing the investment of labour, time, and money. Among the top 181 compounds predicted through virtual screening to be capable of binding to NOR or LasR of P. aeruginosa, we could detect in vivo anti-P. aeruginosa activity in 19 (i.e. 10.4% of all compounds tested) of them in the model host C. elegans. As per our search on PubChem on 1 June 2023, these 19 compounds have yet not been reported to possess any kind of biological activity, and hence we believe this to be the first report of anti-pathogenic activity in these compounds. Further investigation on these active compounds with respect to their mode of action is warranted, which besides confirming their antibacterial activity will also provide additional validation to the targetability of NOR and LasR.
Limitations
The anti-virulence assay performed in this study is not specific to LasR or NOR, hence precise mode of action of active anti-pathogenic compounds warrants further confirmatory assays. We could not carry out in vitro MIC/MBC assay for active compounds owing to limited quantity at our disposal, hence it was not possible to distinguish between growth-inhibitory and virulence-inhibitory (i.e. non-antibiotic action) activity. Further, anti-pathogenic activity of some of the compounds might be masked by their anthelmintic activity (Tabs. II-IV).
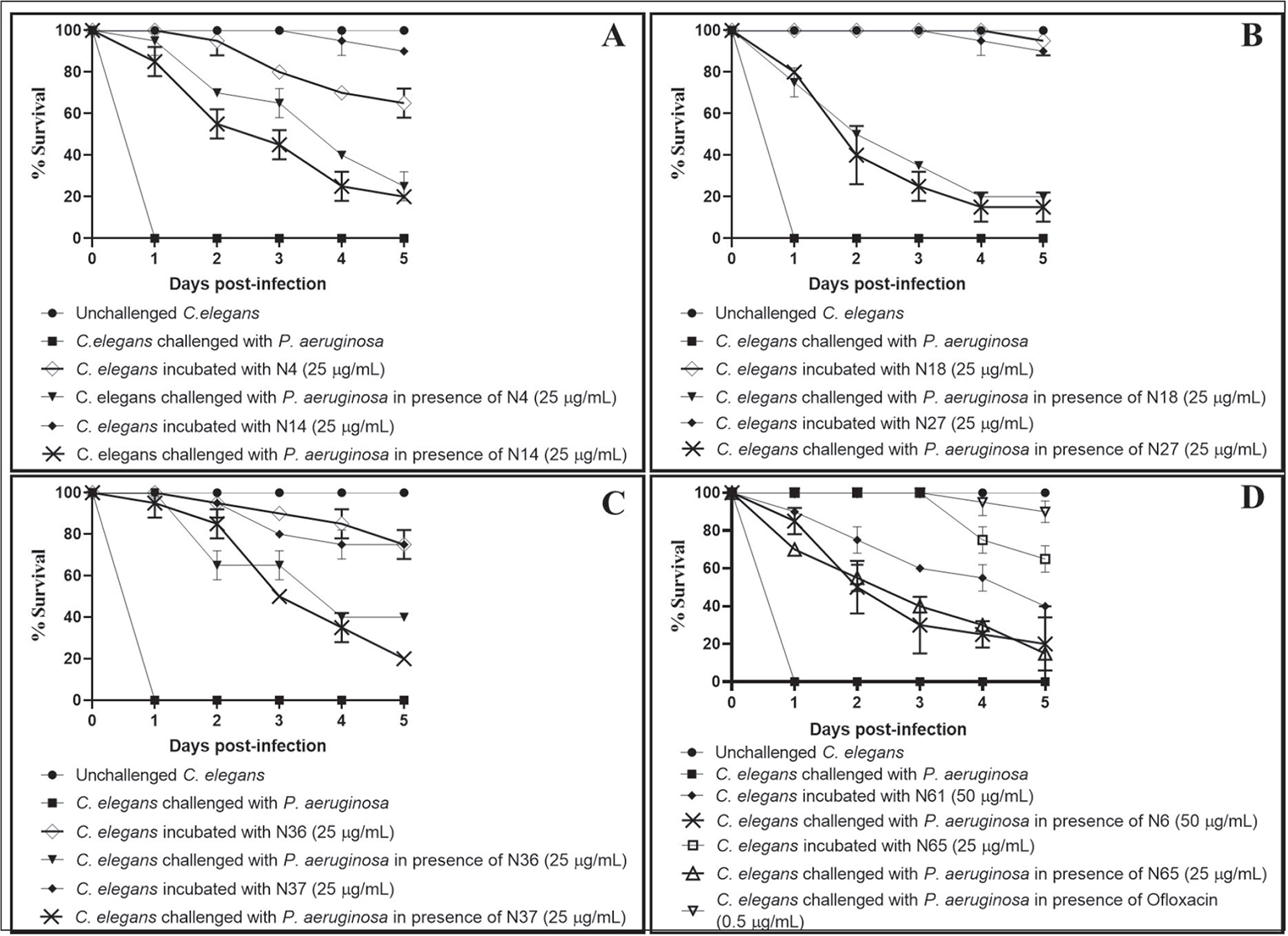
Fig. 4 - Pseudomonas aeruginosa’s virulence towards the host worm gets attenuated in the presence of certain predicted nitric oxide reductase (NOR) inhibitors. A) P. aeruginosa could kill 70%±0%*** and 55%±7%** lesser worms in the presence of N4 (Z954454636) and N14 (Z1765101069), respectively. B) P. aeruginosa could kill 50%±0%*** and 40%±14%** lesser worms in the presence of N18 (Z110018576) and N27 (Z1611882500), respectively. C) P. aeruginosa could kill 65%±7%* and 85%±7%** lesser worms in the presence of N36 (Z397755956) and N37 (Z1190350270), respectively. D) P. aeruginosa could kill 50%±14%**, 55%±0.7%**, and 100%±0%*** lesser worms in the presence of N61 (Z2740017161), N65 (Z1874308288), and ofloxacin (0.5 µg/mL), respectively. Later it was employed as a positive control at its sub-minimum inhibitory concentration (MIC) level, and it did allow progeny formation in worm population from the third day onwards.
Dimethyl sulfoxide (DMSO; 0.5% v/v) present in the ‘vehicle control’ neither affected virulence of the bacterium towards Caenorhabditis elegans, nor did it show any effect on worm survival. **p<0.01, ***p<0.001. The percent values reported pertain to the worm survival 2 days post-infection.
| Lab code | Manufacturer’s code | Conc.(µg/mL) | % reduction in bacterial virulence | |
|---|---|---|---|---|
| 1st day end-point | 2nd day end-point | |||
| N4 | Z954454636 | 25 | 95 ± 7*** | 70 ± 0*** |
| N14 | Z1765101069 | 25 | 85 ± 7** | 55 ± 7** |
| N18 | Z110018576 | 25 | 75 ± 7** | 50 ± 0*** |
| N27 | Z1611882500 | 25 | 80 ± 0*** | 40 ± 14* |
| N36 | Z397755956 | 25 | 100 ± 0*** | 65 ± 7* |
| N37 | Z1190350270 | 25 | 95 ± 7** | 85 ± 7** |
| N61 | Z2740017161 | 50 | 85 ± 7** | 50 ± 14** |
| N65 | Z1874308288 | 25 | 70 ± 0*** | 55 ± 0.7** |
**p<0.01, ***p<0.001.
NOR = nitric oxide reductase.
| Lab code | Manufacturer’s code | Conc (µg/mL) | % anti-pathogenic activity based on fifth day end-point | |
|---|---|---|---|---|
| Without nullifying compound’s toxicity towards worms
(A) |
After nullifying compound’s toxicity towards worms
(B) |
|||
| N4 | Z954454636 | 25 | 25 ± 7** | 60 ± 7** |
| N36 | Z397755956 | 25 | 40 ± 0** | 65 ± 0*** |
| N37 | Z1190350270 | 25 | 20 ± 0*** | 45 ± 0*** |
| N61 | Z2740017161 | 50 | 20 ± 14 | 80 ± 14** |
| N65 | Z1874308288 | 25 | 15 ± 21 | 40 ± 21** |
**p<0.01, ***p<0.001.
NOR = nitric oxide reductase.
Acknowledgements
Authors thank Nirma Education and Research Foundation (NERF), Ahmedabad, for infrastructural support. GG acknowledges scholarship from the Government of Gujarat through their SHODH scheme.
Disclosures
Conflict of interest: The authors declare no conflict of interest.
Financial support: This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Authors contribution: GG: Carried out experiments; participated in manuscript preparation; literature survey; NH and BC: Executed in silico part of the study; arranged procurement of test compounds in physical form; VK: Conceptualization, data analysis, and manuscript preparation
References
- 1. Murray CJ, Ikuta KS, Sharara F, et al; Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629-655. CrossRef PubMed
- 2. Tacconelli E, Carrara E, Savoldi A, et al; WHO Pathogens Priority List Working Group. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318-327. CrossRef PubMed
- 3. Singh SB, Barrett JF. Empirical antibacterial drug discovery – foundation in natural products. Biochem Pharmacol. 2006;71(7):1006-1015. CrossRef PubMed
- 4. Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6(1):29-40. CrossRef PubMed
- 5. Jaeger T, Flohé L. The thiol-based redox networks of pathogens: unexploited targets in the search for new drugs. Biofactors. 2006;27(1-4):109-120. CrossRef PubMed
- 6. Ruparel FJ, Shah SK, Patel JH, Thakkar NR, Gajera GN, Kothari VO. Network analysis for identifying potential anti-virulence targets from whole transcriptome of Pseudomonas aeruginosa and Staphylococcus aureus exposed to certain anti-pathogenic polyherbal formulations. Drug Target Insights. 2023;17:58-69. CrossRef PubMed
- 7. Groleau MC, de Oliveira Pereira T, Dekimpe V, Déziel E. PqsE is essential for RhlR-dependent quorum sensing regulation in Pseudomonas aeruginosa. mSystems. 2020;5(3):10-128. CrossRef PubMed
- 8. Joshi C, Kothari V. Bacterial stress-response machinery as a target for next-generation antimicrobials. Infect Disord Drug Targets. 2022;22(6):e210322202493. CrossRef PubMed
- 9. Porcheron G, Dozois CM. Interplay between iron homeostasis and virulence: fur and RyhB as major regulators of bacterial pathogenicity. Vet Microbiol. 2015;179(1-2):2-14. CrossRef PubMed
- 10. Hofmann L, Hirsch M, Ruthstein S. Advances in understanding of the copper homeostasis in Pseudomonas aeruginosa. Int J Mol Sci. 2021;22(4):2050. CrossRef PubMed
- 11. Chen H, Han J, Wang L. Diels-Alder cycloadditions of N-arylpyrroles via aryne intermediates using diaryliodonium salts. Beilstein J Org Chem. 2018;14(1):354-363. CrossRef PubMed
- 12. Kamal AA, Maurer CK, Allegretta G, Haupenthal J, Empting M, Hartmann RW. Quorum sensing inhibitors as pathoblockers for Pseudomonas aeruginosa infections: a new concept in anti-infective drug discovery. Antibacterials. 2018;II:185-210. CrossRef
- 13. Bonvicini F, Mandrone M, Cosa S. Editorial: pathoblockers and antivirulence agents of plant-origin for the management of multidrug resistant pathogens. Front Microbiol. 2023;14:1201495. CrossRef PubMed
- 14. Huang Y, Chen Y, Zhang LH. The roles of microbial cell-cell chemical communication systems in the modulation of antimicrobial resistance. Antibiotics (Basel). 2020;9(11):779. CrossRef PubMed
- 15. Nandi S. Recent advances in ligand and structure based screening of potent quorum sensing inhibitors against antibiotic resistance induced bacterial virulence. Recent Pat Biotechnol. 2016;10(2):195-216. CrossRef PubMed
- 16. Kumar M, Saxena M, Saxena AK, Nandi S. Recent breakthroughs in various antimicrobial resistance induced quorum sensing biosynthetic pathway mediated targets and design of their inhibitors. Comb Chem High Throughput Screen. 2020;23(6):458-476. CrossRef PubMed
- 17. Abisado RG, Benomar S, Klaus JR, Dandekar AA, Chandler JR. Bacterial quorum sensing and microbial community interactions. MBio. 2018;9(3):10-128. CrossRef PubMed
- 18. Della Sala G, Teta R, Esposito G, Costantino V. The chemical language of gram-negative bacteria. InQuorum Sensing. Academic Press; 2019:3-28. CrossRef.
- 19. Kanak KR, Dass RS, Pan A. Anti-quorum sensing potential of selenium nanoparticles against LasI/R, RhlI/R, and PQS/MvfR in Pseudomonas aeruginosa: a molecular docking approach. Front Mol Biosci. 2023 Aug 10;10:1203672. CrossRef PubMed
- 20. Kostylev M, Kim DY, Smalley NE, Salukhe I, Greenberg EP, Dandekar AA. Evolution of the Pseudomonas aeruginosa quorum-sensing hierarchy. Proc Natl Acad Sci USA. 2019;116(14):7027-7032. CrossRef PubMed
- 21. Schuster M, Urbanowski ML, Greenberg EP. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc Natl Acad Sci USA. 2004;101(45):15833-15839. CrossRef PubMed
- 22. Maddocks SE. Novel targets of antimicrobial therapies. Microbiol Spectr. 2016;4(2):10-128. CrossRef PubMed
- 23. Haque S, Ahmad F, Dar SA, et al. Developments in strategies for Quorum Sensing virulence factor inhibition to combat bacterial drug resistance. Microb Pathog. 2018;121:293-302. CrossRef PubMed
- 24. Geske GD, O’Neill JC, Blackwell HE. Expanding dialogues: from natural autoinducers to non-natural analogues that modulate quorum sensing in Gram-negative bacteria. Chem Soc Rev. 2008;37(7):1432-1447. CrossRef PubMed
- 25. Packiavathy IASV, Kannappan A, Thiyagarajan S, et al. AHL-Lactonase producing Psychrobacter sp. from Palk Bay sediment mitigates quorum sensing-mediated virulence production in Gram negative bacterial pathogens. Front Microbiol. 2021;12:634593. CrossRef PubMed
- 26. Poole K. Stress responses as determinants of antimicrobial resistance in Pseudomonas aeruginosa: multidrug efflux and more. Can J Microbiol. 2014;60(12):783-791. CrossRef PubMed
- 27. Joshi C, Patel P, Palep H, Kothari V. Validation of the anti-infective potential of a polyherbal ‘Panchvalkal’ preparation, and elucidation of the molecular basis underlining its efficacy against Pseudomonas aeruginosa. BMC Complement Altern Med. 2019;19(1):1-5. CrossRef PubMed
- 28. Van Alst NE, Picardo KF, Iglewski BH, Haidaris CG. Nitrate sensing and metabolism modulate motility, biofilm formation, and virulence in Pseudomonas aeruginosa. Infect Immun. 2007;75(8):3780-3790. CrossRef PubMed
- 29. Han S, Liu J, Li M, et al. DNA Methyltransferase regulates nitric oxide homeostasis and virulence in a chronically adapted Pseudomonas aeruginosa strain. mSystems. 2022;7(5):e0043422. CrossRef PubMed
- 30. Toyofuku M, Yoon SS. Nitric oxide, an old molecule with noble functions in Pseudomonas aeruginosa biology. Adv Microb Physiol. 2018;72:117-145. CrossRef PubMed
- 31. Barraud N, Kelso MJ, Rice SA, Kjelleberg S. Nitric oxide: a key mediator of biofilm dispersal with applications in infectious diseases. Curr Pharm Des. 2015;21(1):31-42. CrossRef PubMed
- 32. Carvalho SM, Beas JZ, Videira MAM, Saraiva LM. Defenses of multidrug resistant pathogens against reactive nitrogen species produced in infected hosts. Adv Microb Physiol. 2022;80:85-155. CrossRef PubMed
- 33. Wang Z, Xie X, Shang D, et al. A c-di-GMP signaling cascade controls motility, biofilm formation, and virulence in Burkholderia thailandensis. Appl Environ Microbiol. 2022;88(7):e0252921. CrossRef PubMed
- 34. Kakishima K, Shiratsuchi A, Taoka A, Nakanishi Y, Fukumori Y. Participation of nitric oxide reductase in survival of Pseudomonas aeruginosa in LPS-activated macrophages. Biochem Biophys Res Commun. 2007;355(2):587-591. CrossRef PubMed
- 35. Abelyan N, Grabski H, Tiratsuyan S. In silico screening of flavones and its derivatives as potential inhibitors of quorum-sensing regulator LasR of Pseudomonas aeruginosa. Mol Biol (Mosk). 2020;54(1):153-163. CrossRef PubMed
- 36. Narayanaswamy R, Prabhakaran VS, Al-Ansari MM, Al-Humaid LA, Tiwari P. An in silico analysis of synthetic and natural compounds as inhibitors of nitrous oxide reductase (N2OR) and nitrite reductase (NIR). Toxics. 2023;11(8):660. CrossRef PubMed
- 37. Wallach I, Dzamba M, Heifets A. AtomNet: a deep convolutional neural network for bioactivity prediction in structure-based drug discovery. arXiv preprint arXiv:1510.02855. 2015. CrossRef
- 38. Patel P, Joshi C, Kothari V. Antipathogenic potential of a polyherbal wound-care formulation (herboheal) against certain wound-infective gram-negative bacteria. Adv Pharmacol Sci. 2019;2019:1739868. CrossRef PubMed
- 39. Abd El-Aleam RH, Sayed AM, Taha MN, George RF, Georgey HH, Abdel-Rahman HM. Design and synthesis of novel benzimidazole derivatives as potential Pseudomonas aeruginosa anti-biofilm agents inhibiting LasR: evidence from comprehensive molecular dynamics simulation and in vitro investigation. Eur J Med Chem. 2022;241:114629. CrossRef PubMed
- 40. Hernando-Amado S, Alcalde-Rico M, Gil-Gil T, Valverde JR, Martínez JL. Naringenin inhibition of the Pseudomonas aeruginosa quorum sensing response is based on its time-dependent competition with N-(3-Oxo-dodecanoyl)-L-homoserine lactone for LasR binding. Front Mol Biosci. 2020;7:25. CrossRef PubMed
- 41. O’Brien KT, Noto JG, Nichols-O’Neill L, Perez LJ. Potent irreversible inhibitors of LasR quorum sensing in Pseudomonas aeruginosa. ACS Med Chem Lett. 2015;6(2):162-167. CrossRef PubMed
- 42. Walsh C. Where will new antibiotics come from? Nat Rev Microbiol. 2003;1(1):65-70. CrossRef PubMed
- 43. Allen RC, Popat R, Diggle SP, Brown SP. Targeting virulence: can we make evolution-proof drugs? Nat Rev Microbiol. 2014;12(4):300-308. CrossRef PubMed