 |
Drug Target Insights 2023; 17: 78-89 ISSN 1177-3928 | DOI: 10.33393/dti.2023.2596 REVIEW |
 |
Efficacy of LAMP assay for Mycobacterial spp. detection to prevent treatment delays and onset of drug resistance: a systematic review and meta-analysis
ABSTRACT
Background: Tuberculosis (TB) remains a deadly disease affecting one-third population globally. Long turnaround time and poor sensitivity of the conventional diagnostics are the major impediments for faster diagnosis of Mycobacterial spp to prevent drug resistance. To overcome these issues, molecular diagnostics have been developed. They offer enhanced sensitivity but require sophisticated infrastructure, skilled manpower and remain expensive.
Methods: In that context, loop-mediated isothermal amplification (LAMP) assay, recommended by the WHO in 2016 for TB diagnosis, sounds as a promising alternative that facilitates visual read outs. Therefore, the aim of the present study is to conduct a meta-analysis to assess the diagnostic efficiency of LAMP for the detection of a panel of Mycobacterium spp. following PRISMA guidelines using scientific databases. From 1600 studies reported on the diagnosis of Mycobacterium spp., a selection of 30 articles were identified as eligible to meet the criteria of LAMP based diagnosis.
Results: It was found that most of the studies were conducted in high disease burden nations such as India, Thailand, and Japan with sputum as the most common specimen to be used for LAMP assay. Furthermore, IS6110 gene and fluorescence-based detections ranked as the most used target and method respectively. The accuracy and precision rates mostly varied between 79.2% to 99.3% and 73.9% to 100%, respectively. Lastly, a quality assessment based on QUADAS-2 of bias and applicability was conducted.
Conclusion: LAMP technology could be considered as a feasible alternative to current diagnostics considering high burden for rapid testing in low resource regions.
Keywords: Diagnosis, LAMP, Meta-analysis, Mycobacteria, Therapeutics, Tuberculosis
Received: May 1, 2023
Accepted: May 12, 2023
Published online: June 7, 2023
Corresponding authors:
Drs. Saif Hameed and Zeeshan Fatima
Amity Institute of Biotechnology
Amity University Haryana
Gurugram, Manesar-122413 - India
saifhameed@yahoo.co.in; drzeeshanfatima@gmail.com
Drug Target Insights - ISSN 1177-3928 - www.aboutscience.eu/dti
© 2023 The Authors. This article is published by AboutScience and licensed under Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0).
Commercial use is not permitted and is subject to Publisher’s permissions. Full information is available at www.aboutscience.eu
Introduction
Tuberculosis (TB) caused by Mycobacterium tuberculosis (MTB) remains a deadly disease affecting millions of people worldwide. It is estimated to affect approximately one-third of the global population and is becoming one of the most fatal infectious diseases. MTB usually attacks the lungs, but TB bacteria can infect any part of the body such as kidney, spine, or brain (1). Worldwide, TB is the 13th leading cause of death and the second raging infectious killer after human immunodeficiency virus (HIV)/acquired immunodeficiency syndrome (AIDS) (2). In 2020, an estimated 10 million people got ill with TB worldwide, the infection being divided as 5.6 million men, 3.3 million women, and 1.1 million children. TB affects most of the countries among all age groups and can be fatal if not treated properly. Moreover, the emergence of drug-resistant strains has further complicated the problem and has become a rising obstacle against efficient therapeutics (3). Therapeutics are available but the effective control of the disease is impeded due to the lack of rapid and accurate diagnostics. Under such significant circumstances, there is an urgent need for rapid, accurate, and cost-effective diagnostic test for TB to identify new cases and reduce the time-to-treatment and prevent its further transmission.
The current available methods are primarily based on smear microscopy (acid-fast staining), culture, and nucleic acid amplification. Although methods based on acid-fast staining are sensitive, they pose problems in low-resource places and are time-consuming (4). The solid culture method requires around 4-8 weeks, while liquid-based culture methods also require around 10-14 days (4). Nucleic acid amplification techniques are based on polymerase chain reaction (PCR) or loop-mediated isothermal amplification (LAMP). Although hemi-nested PCR based on GeneXpert for MTB detection is rapid, sensitive, and specific, it also poses challenges of high cost and high end equipment dependency, which limits its implementation in low-resource regions (5). LAMP is an isothermal DNA amplification method that relies on four or six pairs of primers to amplify minute quantities of DNA within a shorter period with simple operation, making it more suitable for low-resource regions (6). Thus, research in TB diagnostics aims to find an efficient, reproducible, cost-effective tool with minimal infrastructure requirements. LAMP is a popularly adopted new age technology for rapid nucleic acid amplification which is widely used for pathogen (virus, bacteria, and malaria) detection including severe acute respiratory syndrome coronavirus 2 (SARS CoV-2) (7-9). LAMP-based detection methods have been proved to be more sensitive than GeneXpert assay. In fact, the World Health Organization (WHO) has endorsed LAMP for TB as a replacement for smear microscopy for peripheral settings (10).
In pursuit of developing better diagnostics, which are crucial for achieving global elimination of TB, we performed a systematic review and meta-analysis to access the diagnostic accuracy of LAMP to detect mycobacteria. Even if couple of studies have depicted the efficacy of LAMP during the last decade, an updated version is missing. Moreover, most of these studies were specific to either pulmonary or extrapulmonary TB. Therefore, the present study not only offers an up-to-date diagnostic performance of LAMP for TB detection but also covers other Mycobacterium spp. The pooled sensitivity and specificity of LAMP were analyzed against different references. Further, diagnostic efficiency was determined based on reference methods, target genes, and detection methods of LAMP. Taken together, we aimed to evaluate the diagnostic potency of LAMP as a tool for detection of mycobacteria to address the current TB diagnosis burden in low-resource places.
Methods
The Preferred Reporting Items for Systematic Review and Meta-Analyses (PRISMA) guidelines (11) were followed for identification of eligible studies in the present systematic review and meta-analysis.
Search strategy
Diverse scientific databases, for example, PubMed, Google Scholar, Science Direct, Scopus, BioRxiv, and MedRxiv, were searched to screen for studies performed using LAMP for TB diagnostics from the year 2000 till March 2022. The terms such as LAMP, Tuberculosis, Mycobacterium and mycobacteria were used in various combinations during our research without any limitations: “LAMP + Tuberculosis” or “LAMP + Mycobacterium” or “LAMP + mycobacteria” or “LAMP + TB” or “LAMP + Tuberculosis + Mycobacterium” or “LAMP + Tuberculosis + mycobacteria” for PubMed, Science Direct, and Google Scholar without using any language restriction. The retrieved results were screened for duplication and conformity with the prespecified eligibility criteria.
Study eligibility criteria
Inclusion criteria
This systematic review and meta-analysis included: (1) both peer-reviewed and preprint original articles on LAMP technology used for detection of any mycobacterial species such as MTB, M. bovis, and M. africanum; (2) only full-text articles written in English language; and (3) articles that contain data on true-positive (TP), false-positive (FP), false-negative (FN), and true-negative (TN) values for the assay or have sufficient data so that the number of TP, FP, FN, and TN (performed on clinical samples) could be determined.
Exclusion criteria
Exclusion was made for: (1) studies based on non-isothermal amplification; (2) studies where data are irretrievable; (3) review articles, editorials, commentaries, proceedings, etc.; (4) foreign language articles (other than English) based on LAMP-mediated detection of mycobacteria.
Data extraction
Potential articles after reviewing titles and abstracts followed by full text for inclusion were extracted by two authors (G.S.B. and Z.H.). Consultation from two independent authors (S.J. and S.H.) was made to eliminate the doubt about any discrepancy. The extracted information from included studies had authors, year of publication, location of study, sample size, types of specimens, target genes, detection method, and standard reference method. The data extracted for evaluation of diagnostic accuracy for LAMP were performed by using either respiratory or non-respiratory specimens with any of the reference methods such as smear microscopy, culture, and GeneXpert. The important parameters in this meta-analysis such as TP, TN, FP, and FN of all studies were either extracted or calculated to provide their sensitivity and specificity values. The included studies (n = 30) were then assessed for their methodological quality to reduce systematic biases and inferential errors from the collected data.
Statistical analysis
The quantitative analysis of the included studies (n = 30) from the data extracted such as the values of TP, FP, TN, FN and sample size was performed. Furthermore, the values of sensitivity and sensitivity were mined or calculated from the available data. Moreover, pooled sensitivity and specificity of LAMP associated with 95% confidence interval (CI) were estimated. To maintain the accuracy and precision, the formulas: Accuracy = [TP + TN/TP + TN + FP + FN]* 100 and Precision = [TP/TP+FP]* 100 (12,13) were used. Accuracy and precision are important characteristics of any measurement. Accuracy is the degree of closeness of measured value to a standard value. However, precision provides the information regarding the closeness of multiple measured values to each other. Accuracy and precision are independent of each other. Forest plot for sensitivity and specificity were plotted using R-software along with summary receiver operating characteristic (SROC) for the given study.
Quality assessment
Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) tool was used to assess the methodological quality of the eligible studies. The risk of bias in the included studies (n = 30) was assessed from four areas of bias, for example, patient selection, index test, reference standards, and flow timing (14,15). For each QUADAS-2 domain specific yes/no questions were tailored. Following these criteria, the eligible studies were then refereed for low, unclear, or high risks of bias. Furthermore, we also judged to generate low, unclear, or high-risk applicability.
Results
Literature survey
We followed the PRISMA guidelines (11) to search the literature for the present study (Fig. 1). The major scientific databases viz. PubMed, Science Direct, Scopus, BioRxiv, and MedRxiv have been extensively searched applying the above inclusion criteria and around 1,600 articles were extracted. From the 1,600 articles, we included the ones that were published after the year 2000 since the inception of LAMP technology (6) and thus excluded 22 articles. Further, only articles written in English language were considered and thus excluded 44 articles. Reading the titles and abstracts of these studies allowed to exclude further 1,029 articles comprising the review articles, editorials, proceedings etc. Following this exclusion, we removed the duplicated articles and further excluded 390 articles. Additional 73 articles were irrelevant as they didn’t use LAMP technology for the diagnosis of any mycobacterial species and were excluded, leaving a panel composed of 42 eligible studies. Lastly, from the 42 included articles, further 12 articles were also eliminated because their TP, FP, TN, and TN values were either not specified in these articles or the sensitivity and specificity values could not be calculated. Altogether, we observed that only 30 articles were eligible for detailed meta-analysis (Fig. 1) considering all the exclusion criteria.
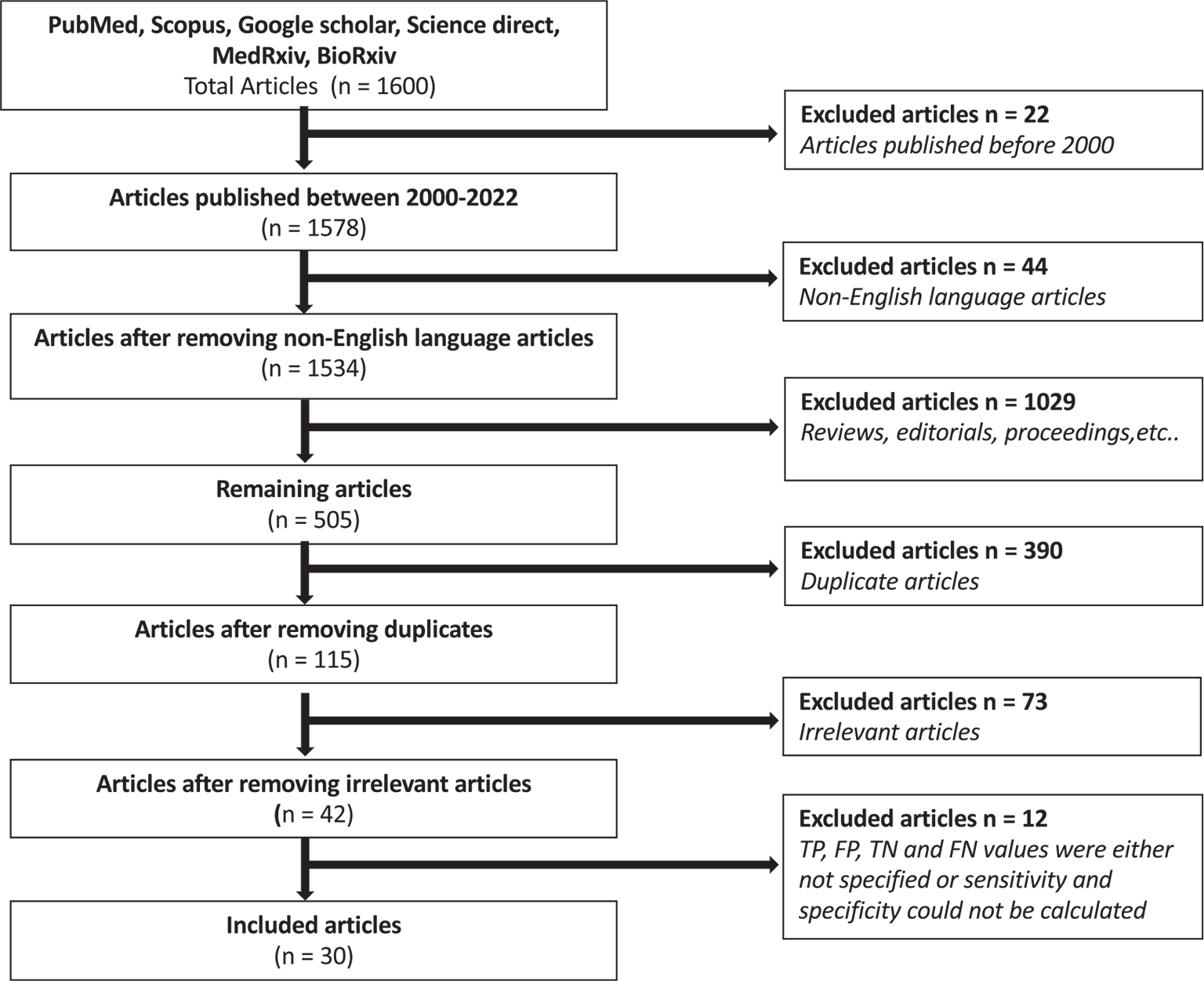
Fig. 1 - Preferred Reporting Items for Systematic Review and Meta-Analyses (PRISMA) flowchart depicts search of the literature and screening strategy for meta-analysis.
Study characteristics and meta-analysis
Table I shows the data extracted from the eligible studies mentioning the details of authors, year of publication, country of study, types of specimens, target genes, detection method, and reference methods. Figure 2 shows the country-wise distribution of 30 identified articles included in the present study. Most of the studies (43.3%; n = 13) were conducted in the high TB burden nations such as India followed by Thailand and Japan (each 10%; n = 3). Two studies each were also conducted in countries such as China, Korea, and Switzerland (6.3%; n = 2). Apart from this, one study each, that is, 3.3%, was from countries included such as Gambia, Nepal, South Africa, Sri Lanka, and Vietnam. Although most of the included articles do not mention about the patient details, the type of specimen (Fig. 3) used in most of the studies was sputum (42.8%; n = 21). In addition, some studies have been tested on other specimens such as cerebrospinal fluid (n = 4), fecal samples (n = 1), urine (n = 3), blood (n = 2), and pleural fluid (n = 3) for the detection of mycobacteria by using LAMP. Furthermore, the standard smear microscopy, culture assay, and PCR-based methods were used as references either alone or in combination (n = 30). Of note, radiology was also used (n = 4) to validate LAMP results as a reference standard (Tab. I), with one study using immunochromatography (16). Next, we examined the various target genes used for the eligible studies. Ten different types of target genes including hspX, IS900, mpt64, Pab, sdaA, rimM, 16SrRNA, MPB64, gyrB, and IS6110 were used in the included studies (n = 30). IS6110 gene was most frequently used in the included studies (n = 14; 31.18%) followed by gyrB (n = 9, 20.45%), 16SrRNA (n = 6, 13.63%), and MPB64 (n = 6, 13.63%) genes (Fig. 4). Furthermore, while analyzing detection methods used for these 30 studies, fluorescent method (n = 19, 32.39%) was the most frequently performed followed by colorimetry (n = 14, 25.35%), gel electrophoresis (n = 11, 20.00%) and turbidity (n = 9, 16.36%) methods (Fig. 5). In 53.33% (n = 16) of studies, more than one detection method was used. In 16.66% (n = 5) of studies, combination of three methods was used while in only two studies (6.66%), combination of four different methods was reported (17,18).
| S. No. | Author | Journal | Country | Reference Method | Specimen | Target Gene | Detection Method | TP | TN | FP | FN | Size | Sensitivity | Specificity |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Boehme et al (2007) | J Clin Microbiol | Switzerland | Culture, smear microscopy | Sputum | gyrB | Fluorescence, turbidity | 173 | 500 | 4 | 5 | 682 | 97.70% | 99% |
| 2 | Pandey et al (2008) | J Med Microbiol | Japan | Acid-fast staining, bacterial culture, radiology | Sputum | 16S rRNA | Fluorescence | 90 | 98 | 6 | 6 | 200 | 94% | 94.20% |
| 3 | Poudel et al (2009) | Kathmandu Univ Med J (KUMJ) | Nepal | Smear microscopy, culture, radiology | Sputum | 16S rRNA | Fluorescence | 97 | 96 | 6 | 3 | 202 | 97% | 94.12% |
| 4 | Geojith et al (2011) | J Microbiol Methods | India | Culture, PCR reverse-hybridization line probe assay, genotype MTBE assay | Sputum | rimM | Colorimetry, gel electrophoresis | 17 | 17 | 1 | 21 | 56 | 44.70% | 94.40% |
| 5 | George et al (2011) | PLoS One | India | Fluorescence smear microscopy, culture | Sputum | rimM | Colorimetry, gel electrophoresis | 31 | 36 | 2 | 2 | 71 | 93.90% | 94.70% |
| 6 | Mitarai et al (2011) | Int J Tuberc Lung Dis | Japan | Culture, smear microscopy, nucleic acid amplification (NAA) | Sputum | gyrB | Fluorescence, turbidity | 196 | 88 | 9 | 27 | 320 | 87.90% | 90.70% |
| 7 | Nagdev et al (2011) | J Clin Microbiol | India | PCR | Cerebrospinal fluid | IS6110 | Turbidity | 15 | 8 | 2 | 2 | 27 | 88.23% | 80% |
| 8 | Sethi et al (2013) | J Clin Lab Anal | India | Smear microscopy, culture, PCR | Sputum | 16S rRNA, IS6110 | Colorimetry, gel electrophoresis | 87 | 30 | 0 | 16 | 133 | 84.50% | 100% |
| 9 | Cao et al (2015) | J Microbiol Methods | China | Smear microscopy, culture, PCR | Sputum | IS6110 | Fluorescence | 98 | 18 | 5 | 2 | 123 | 98.00% | 78.30% |
| 10 | Joon et al (2015) | Int J Tuberc Lung Dis | India | Smear microscopy, culture, PCR | Endometrial fluid, urine, blood, semen, cerebrospinal fluid, pleural fluid, pus, pericardial fluid, peritoneal fluid, intestinal and lymph node biopsy tissue | IS6110, MPB64, sdaA | Colorimetry | 28 | 262 | 23 | 2 | 315 | 93.30% | 91.90% |
| 11 | Moon et al (2015) | J Med Microbiol | Korea | Culture, smear microscopy | Sputum | hspX | Colorimetry, turbidity, gel electrophoresis | 32 | 255 | 3 | 13 | 303 | 71.10% | 98.80% |
| 12 | Bojang et al (2016) | J Infect | Gambia | Smear microscopy, culture, GeneXpert MTB/RIF | Sputum | 16S rRNA | Fluorescence | 98 | 157 | 10 | 1 | 266 | 99.00% | 94.00% |
| 13 | Gray et al (2016) | J Clin Microbiol | Switzerland | Culture, smear microscopy | Sputum | gyrB | Fluorescence | 331 | ### | 52 | 61 | 1777 | 84.40% | 96.60% |
| 14 | Kaku et al (2016) | Jpn J Infect Dis | Japan | Smear microscopy, culture | Sputum | gyrB, IS6110 | Fluorescence | 134 | 312 | 5 | 21 | 472 | 86.50% | 98.40% |
| 15 | Modi et al (2016) | Int J Tuberc Lung Dis | India | Culture, radiology, staining, PCR | Cerebrospinal fluid | IS6110, MPB64 | Fluorescence, gel electrophoresis, turbidity | 144 | 100 | 0 | 6 | 250 | 96.00% | 100.00% |
| 16 | Sharma et al (2016) | Tuberculosis (Edinb) | India | PCR, culture, smear microscopy | Needle aspirate | IS6110, MPB64 | Fluorescence, gel electrophoresis, turbidity | 108 | 50 | 0 | 12 | 170 | 90.00% | 100.00% |
| 17 | Sharma et al (2016) | J Orthop Res | India | Culture, staining, PCR | Synovial fluid, pus | IS6110, MPB64 | Fluorescence, gel electrophoresis, turbidity | 81 | 50 | 0 | 9 | 140 | 90.00% | 100.00% |
| 18 | Joon et al (2017) | J Microbiol Methods | India | PCR, culture, smear microscopy | Sputum | IS6110, MPB64 | Colorimetry, gel electrophoresis | 17 | 212 | 6 | 1 | 236 | 94.40% | 97.20% |
| 19 | Reddy et al (2017) | Int J Tuberc Lung Dis | South Africa | Culture, smear microscopy, Xpert | Sputum | gyrB | Fluorescence | 119 | 514 | 17 | 45 | 695 | 72.60% | 96.80% |
| 20 | Yadav et al (2017) | Int J Tuberc Lung Dis | India | Culture, smear microscopy, GeneXpert | Sputum | gyrB, IS6110 | Fluorescence | 82 | 368 | 3 | 0 | 453 | 100.00% | 99.20% |
| 21 | Kim et al (2018) | Ann Lab Med | Korea | Culture, smear microscopy, PCR | Sputum | gyrB, IS6110 | Fluorescence, turbidity | 87 | 186 | 0 | 17 | 290 | 83.60% | 100.00% |
| 22 | Nguyen et al (2018) | Diagn Microbiol Infect Dis | Vietnam | Smear microscopy, culture, Xpert MTB/RIF | Sputum | gyrB, IS6110 | Colorimetry, fluorescence | 15 | 445 | 23 | 18 | 501 | 45.50% | 95.10% |
| 23 | Perera et al (2018) | Ceylon Med J | Sri Lanka | Smear microscopy, culture | Culture isolates | rimM | Colorimetry | 31 | 10 | 5 | 0 | 46 | 100.00% | 66.67% |
| 24 | Joon et al (2019) | J Microbiol Methods | India | Culture, smear microscopy, GeneXpert MTB/RIF assay, PCR, LAMP-LFD assay | Sputum | sdaA | Colorimetry | 13 | 92 | 2 | 0 | 107 | 100.00% | 97.87% |
| 25 | Phetsuksiri et al (2019) | Jpn J Infect Dis | Thailand | Culture, immunochromatographic test | Sputum | mpt64 | Colorimetry | 144 | 5 | 1 | 1 | 151 | 99.31% | 83.33% |
| 26 | Punati et al (2019) | Braz J Microbiol | India | Culture, PCR | Fecal samples | IS900 | Turbidity, gel electrophoresis, colorimetry, lateral flow device | 86 | 294 | 9 | 0 | 389 | 100.00% | 97.02% |
| 27 | Rajput et al (2019) | J Microbiol Methods | India | Culture, smear microscopy, PCR | Fluids, urine, pus | IS6110, Pab, MPB64 | Colorimetry, fluorescence | 90 | 32 | 9 | 23 | 154 | 79.65% | 78.05% |
| 28 | Han et al (2020) | BMC Infect Dis | China | Xpert MTB/RIF, SAT-TB assay | Pleural fluids | gyrB, IS6110 | Fluorescence | 59 | 41 | 1 | 164 | 265 | 26.50% | 97.60% |
| 29 | Phetsuksiri et al (2020) | Jpn J Infect Dis | Thailand | Microscopy, culture, PCR, radiology | Sputum | 16S rRNA | Colorimetry, fluorescence, gel electrophoresis, immunochromatography | 119 | 102 | 24 | 7 | 252 | 94.44% | 80.95% |
| 30 | Phetsuksiri et al (2020) | Rev Inst Med Trop Sao Paulo | Thailand | Xpert MTB/RIF, culture, smear microscopy | Sputum | 16S rRNA | Colorimetry, fluorescence, gel electrophoresis | 126 | 71 | 0 | 7 | 204 | 94.74% | 100.00% |
LAMP = loop-mediated isothermal amplification; LFD = lateral flow dipstick; PCR = polymerase chain reaction.
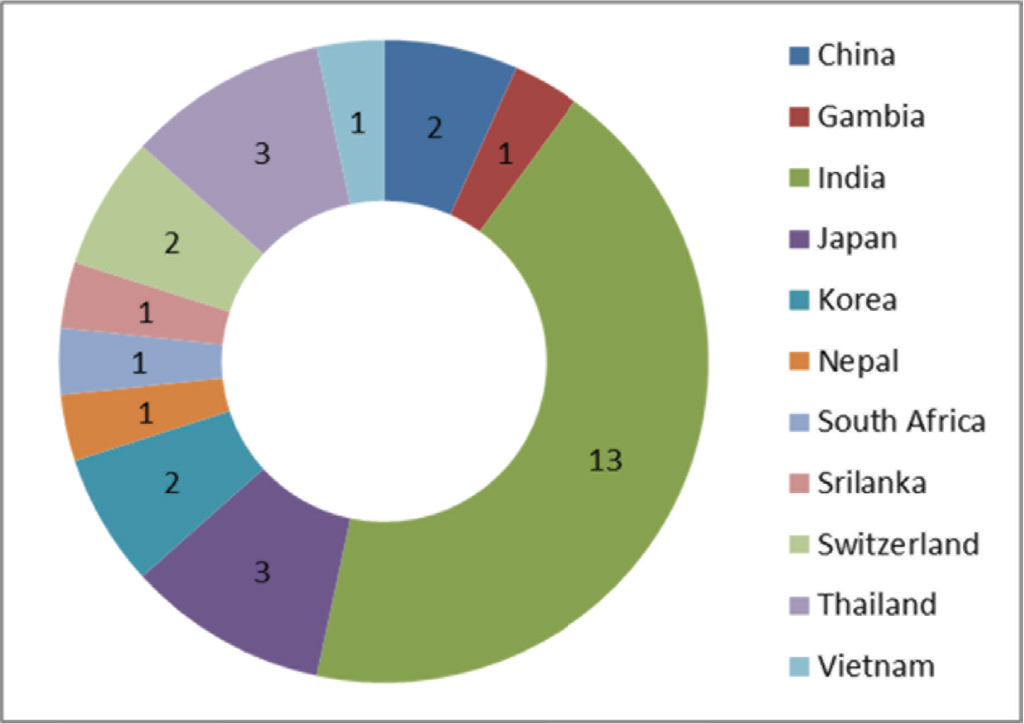
Fig. 2 - Country-wise distribution of included studies (n = 30) reported in the present investigation.
Among all the eligible studies, 4 studies showed 100% sensitivity, while for 16 studies this parameter was higher than 90%. Similarly, 6 studies exhibited 100% specificity while 90% or more specificity was observed in 24 studies (Tab. I). Furthermore, upon analysis of sensitivity and specificity using forest plot at 95% CI, we found that the sensitivity values varied between 0.26 and 1.00 and the specificity values ranged from 0.67 to 1.00 (Fig. 6). A total of 27 out of the 30 included studies showed pooled sensitivity greater than 70%. Only three studies reported sensitivity values of 26% and 45% each (19-21). In terms of FP rate (1-specificity), 27 included studies showed a pooled FP rate higher than 80% (Fig. 7). Additionally, the accuracy and precision rates of included studies were calculated and varied between 37.73% and 99.33%. The analysis proved that 22 studies displayed more than 90% accuracy with only 4 studies depicting less than 80% accuracy (Tab. II). Likewise, the precision rates varied between 39.47% and 100%. The analysis showed that 21 studies exhibited more than 90% precision rate with only 3 studies depicting less than 80%. Of note, we observed that six studies displayed 100% precision rate.
| S. No. | Study | Accuracy | Precision |
|---|---|---|---|
| 1 | Boehme et al (2007) | 98.68 | 97.74 |
| 2 | Pandey et al (2008) | 94.00 | 93.75 |
| 3 | Poudel et al (2009) | 95.54 | 94.17 |
| 4 | Geojith et al (2011) | 60.71 | 94.44 |
| 5 | George et al (2011) | 94.36 | 93.93 |
| 6 | Mitarai et al (2011) | 88.75 | 95.60 |
| 7 | Nagdev et al (2011) | 85.18 | 88.23 |
| 8 | Sethi et al (2013) | 87.96 | 100.00 |
| 9 | Cao et al (2015) | 94.30 | 95.14 |
| 10 | Joon et al (2015) | 92.06 | 54.90 |
| 11 | Moon et al (2015) | 94.71 | 91.42 |
| 12 | Bojang et al (2016) | 95.86 | 90.74 |
| 13 | Gray et al (2016) | 93.64 | 86.42 |
| 14 | Kaku et al (2016) | 94.49 | 96.40 |
| 15 | Modi et al (2016) | 97.60 | 100.00 |
| 16 | Sharma et al (2016) | 92.94 | 100.00 |
| 17 | Joon et al (2017) | 97.03 | 73.91 |
| 18 | Reddy et al (2017) | 91.07 | 87.50 |
| 19 | Sharma et al (2016) | 93.57 | 100.00 |
| 20 | Yadav et al (2017) | 99.33 | 96.47 |
| 21 | Kim et al (2018) | 94.13 | 100.00 |
| 22 | Nguyen et al (2018) | 91.81 | 39.47 |
| 23 | Perera et al (2018) | 89.13 | 86.11 |
| 24 | Joon et al (2019) | 98.13 | 86.66 |
| 25 | Phetsuksiri et al (2019) | 98.67 | 99.31 |
| 26 | Punati et al (2019) | 97.68 | 90.52 |
| 27 | Rajput et al (2019) | 79.22 | 90.90 |
| 28 | Han et al (2020) | 37.73 | 98.33 |
| 29 | Phetsuksiri et al (2020) | 87.69 | 83.21 |
| 30 | Phetsuksiri et al (2020) | 96.56 | 100.00 |
Quality assessment of the study
Almost two-thirds of the included studies (22 out of 30 studies) have a high risk of patient selection bias due to non-random patient selection and case-control study design (Fig. 8, Tab. I). Around 26% (8 out of 30) of the included studies have low risk of patient selection bias because these studies provided sufficient details about patient inclusion/exclusion criteria; 86% of included articles (26 out of 30 studies) present low risk of index test bias because these tests clearly stated the quantitative detection read-outs with reported thresholds. Moreover, these studies explicitly declared that their index and reference tests were done simultaneously in parallel to each other or that testing was blinded from each other. Two studies (19,22) were reported without defined detection thresholds. One study (23) had unclear risk of index test bias as the quantitative detection thresholds were not explained. It was either unclear whether index test results were interpreted with knowledge of reference test results or if only qualitative read-out was used for reading the results. Hence, index test bias of these studies was unclear. For the rest of the included studies, almost all (n = 30) have low risk of reference standard bias because they provided enough information about the standard reference test used in the study.
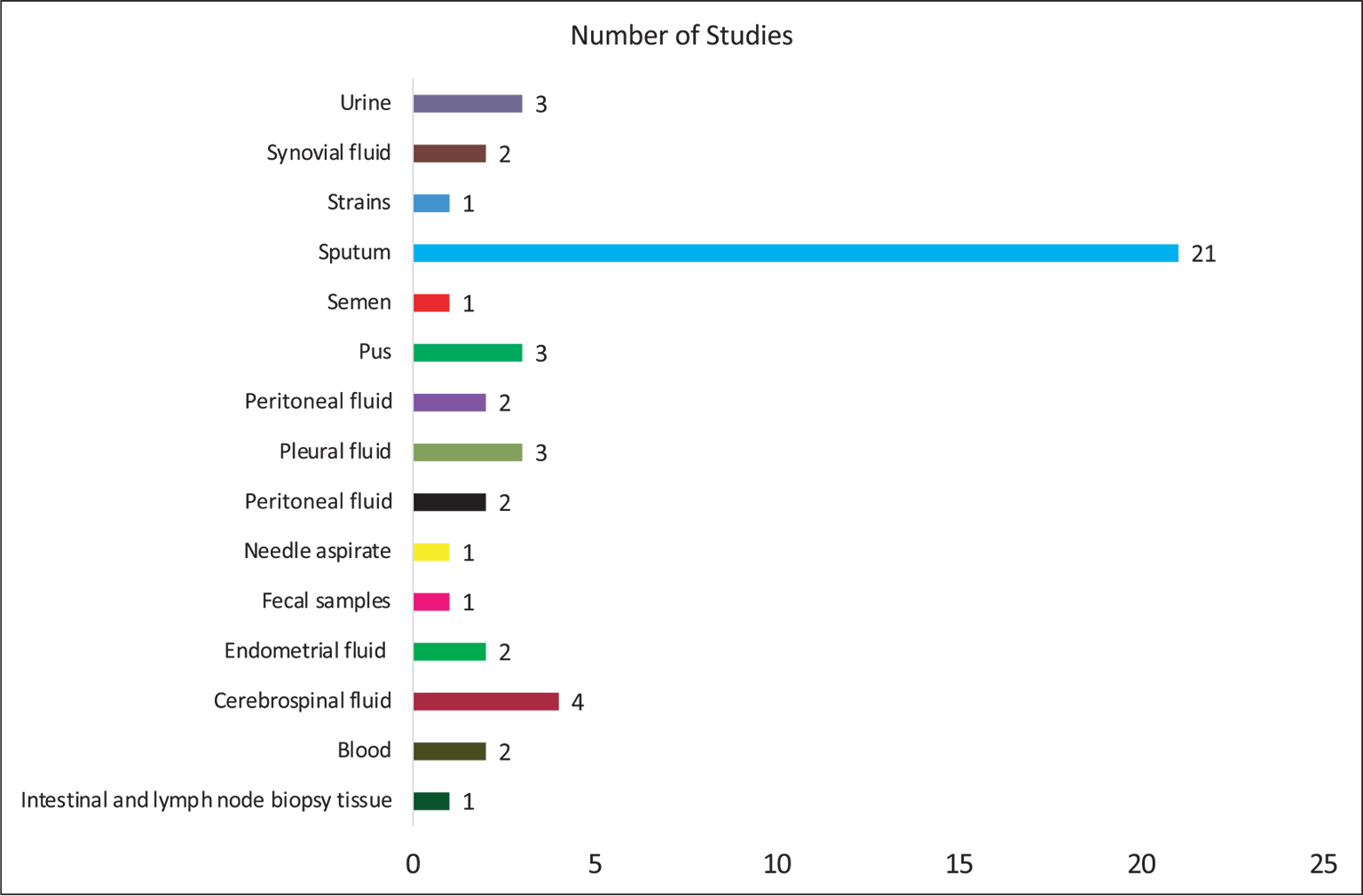
Fig. 3 - Distribution of type of specimen for detection of mycobacteria in the included studies (n = 30).
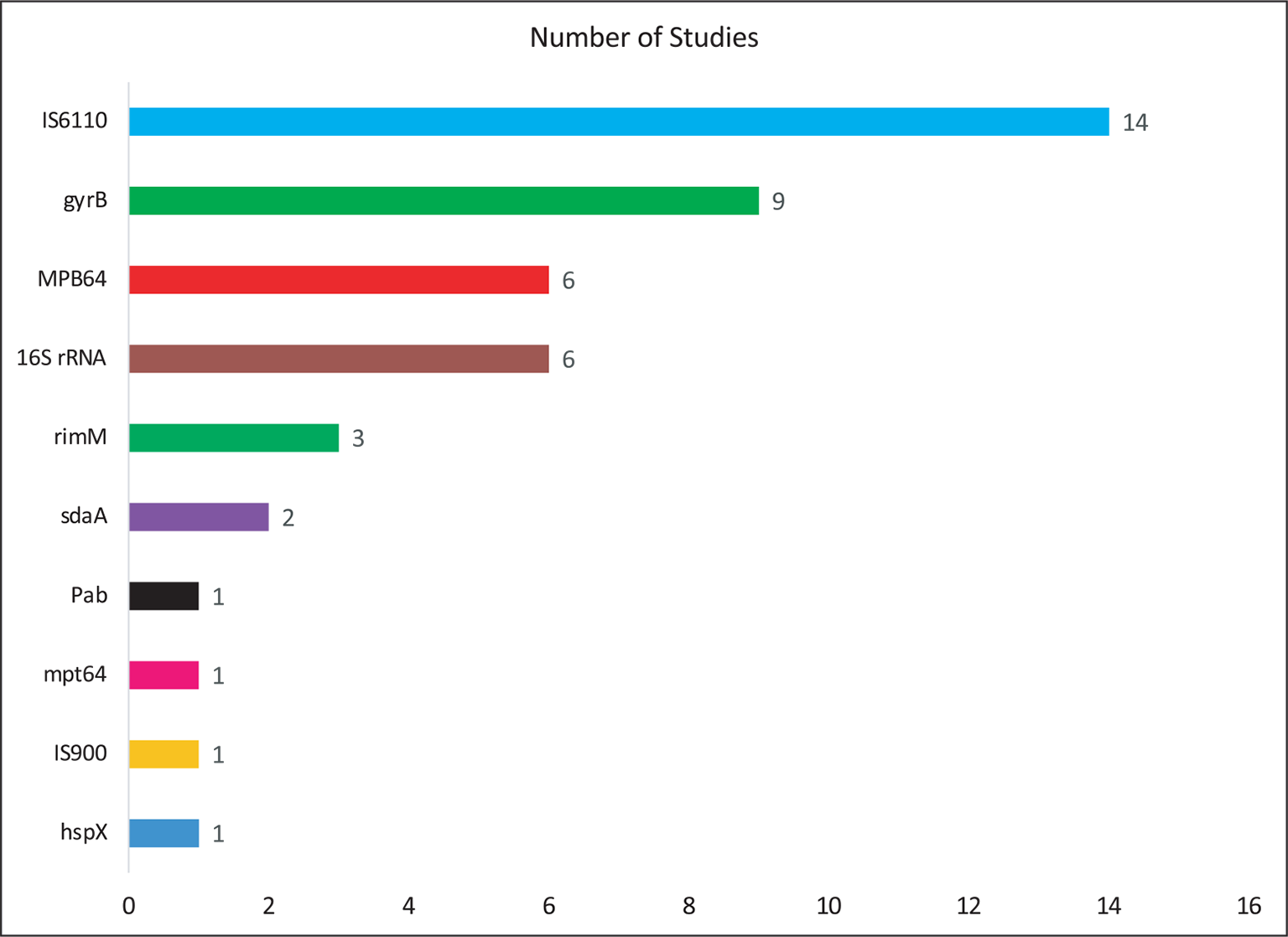
Fig. 4 - Distribution of target genes reported in the included studies (n = 30).
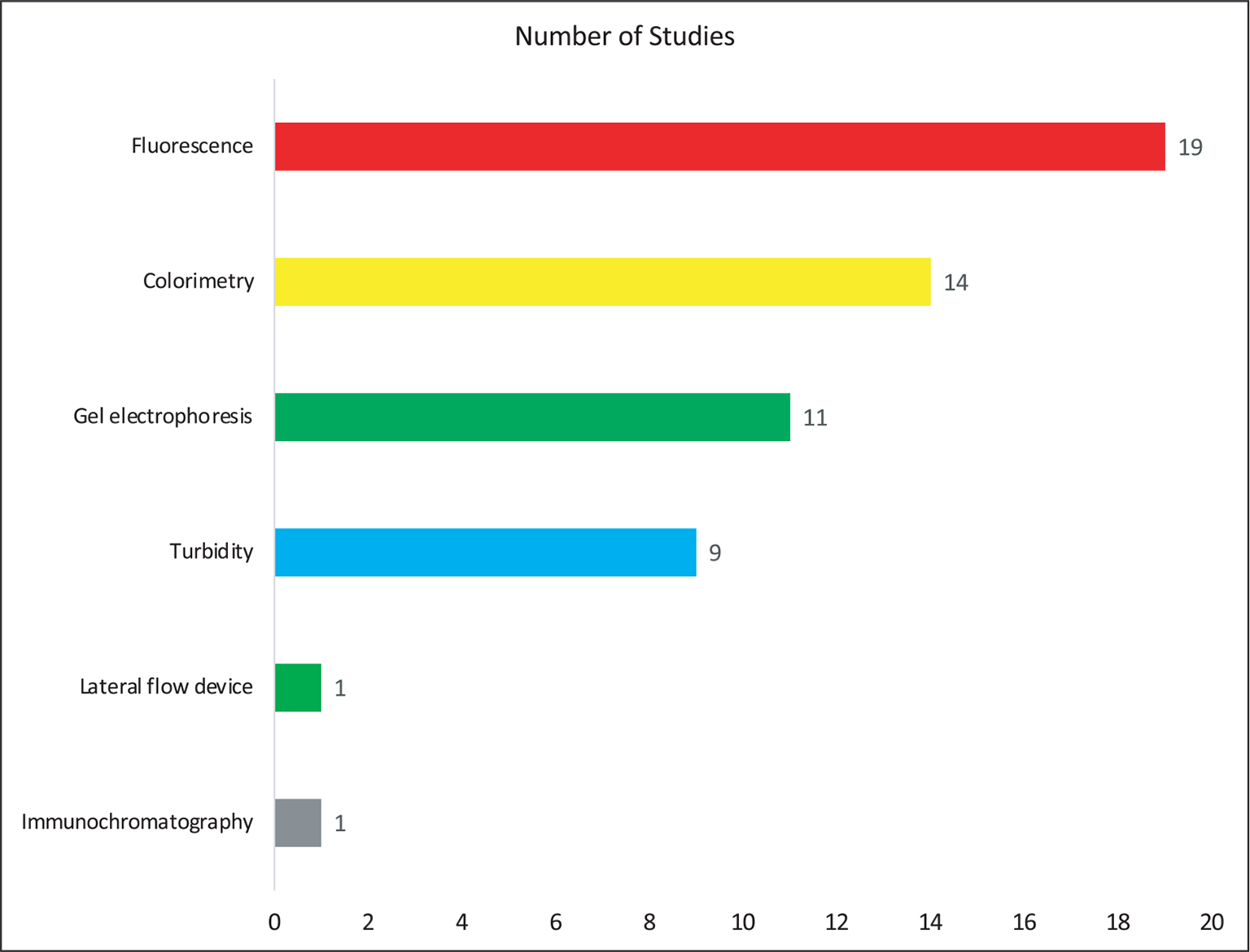
Fig. 5 - Distribution of type of detection method for mycobacteria in the included studies (n = 30).
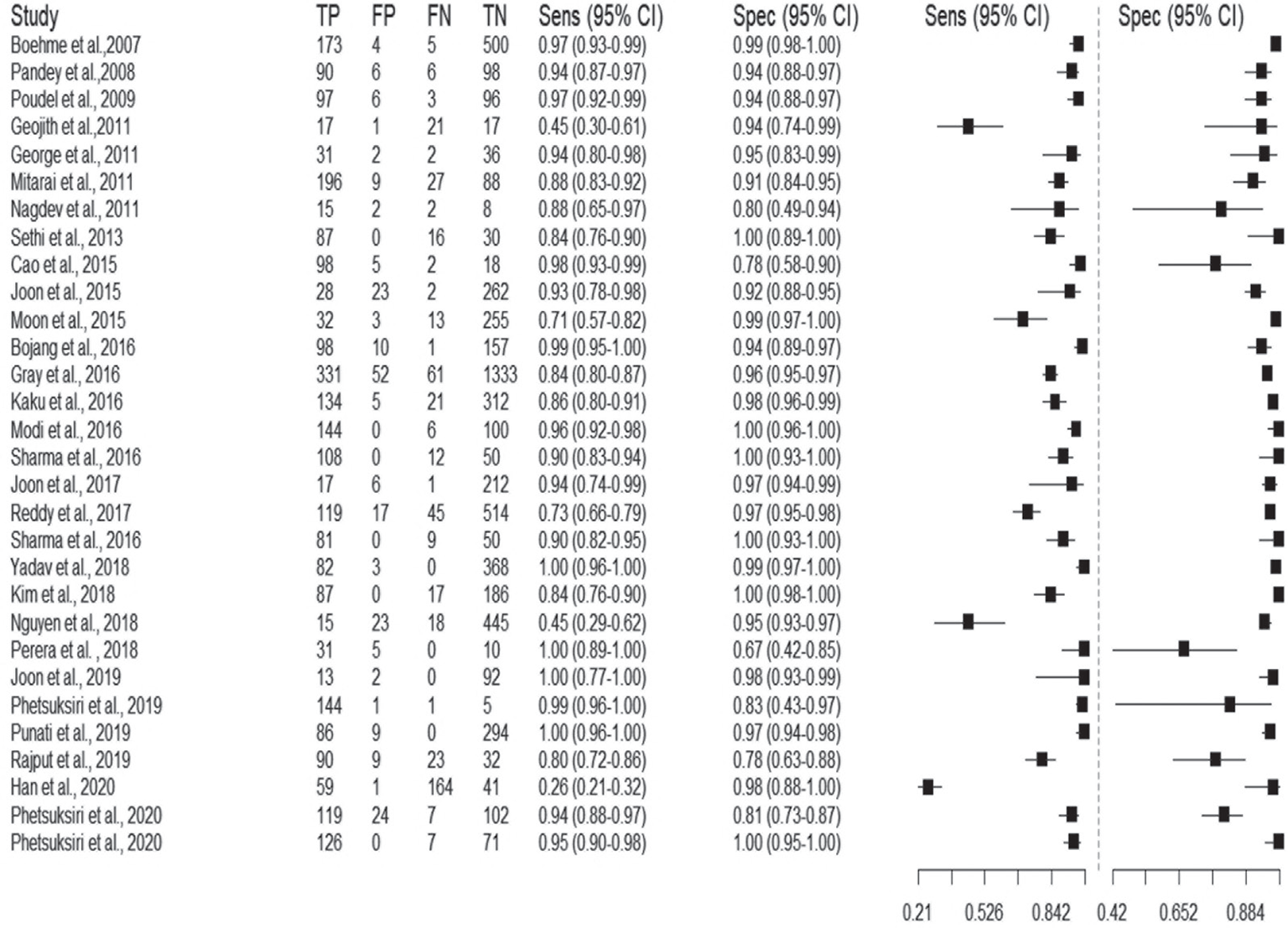
Fig. 6 - The Forest plot of sensitivity and specificity of included studies (n = 30) on the diagnostic performance of loop-mediated isothermal amplification (LAMP) technique.
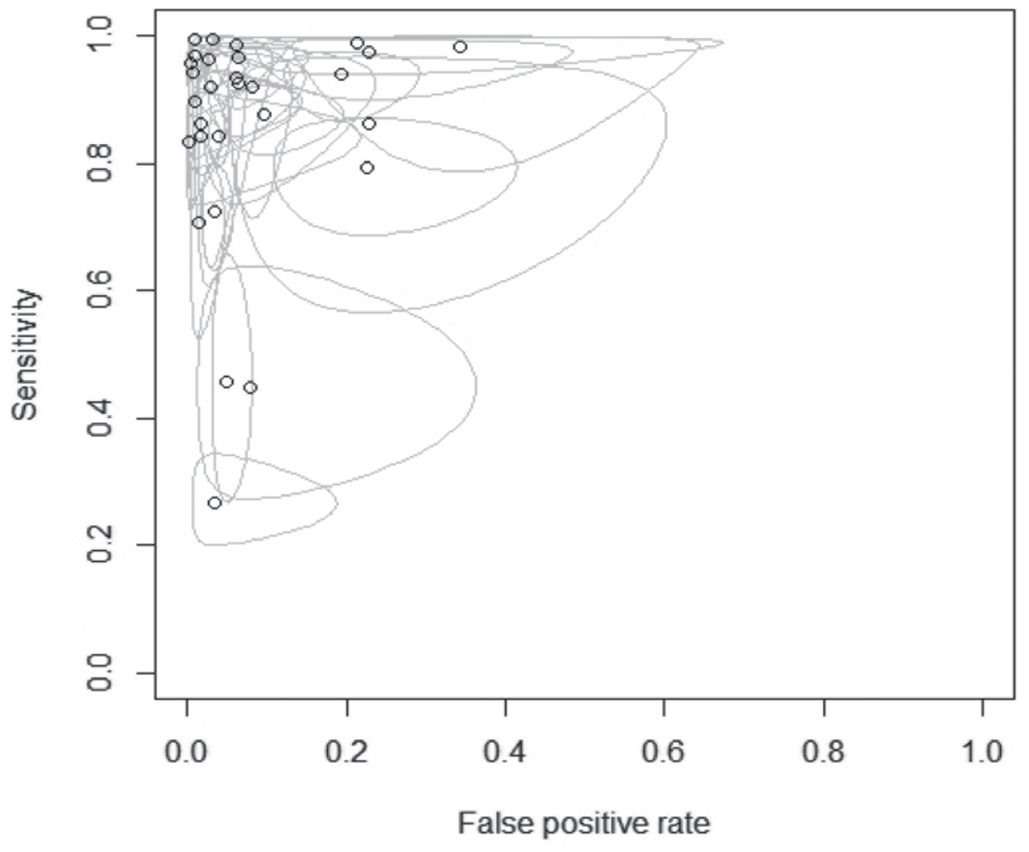
Fig. 7 - Summary receiver operating characteristic (SROC) depicts loop-mediated isothermal amplification (LAMP) diagnostic performance in mycobacteria diagnosis.
Half of the studies (15 out of 30) have an unclear risk of flow and timing bias as there is not enough information, whether reference standard results were interpreted with the knowledge of the results of the index test. One study (24) was at high risk as it did not provide any information on whether the samples for a reference test and the index test were taken at the same time. Our review question did not focus on any patient demographics. None of the included studies attempted to exclude patients based on demographics and thus had no “concern of patient selection applicability” (Fig. 8, Tab. I). Index tests of all studies have generally been used for Point-of-care test (POCTs) and thus have low concern of index test applicability. Reference standard tests of nearly all studies were culture, smear microscopy, Xpert test, PCR, or combinations of them. Thus, we graded these studies as having low concern of standard test applicability.
Discussion
Early and correct diagnosis of all the TB forms is pertinent for effective treatment of the disease and prevention of the spread of infection, particularly in nations which have high burden. The currently available diagnostics rely mostly on smear microscopy, culture, and PCR-based methods which are not only time-consuming and low sensitive but cumbersome and costly (25,26). LAMP assay provides a faster and innovative point-of-care diagnostic alternative as it is cost-effective, sensitive, and gives results in less than 1 hour due to amplification under isothermal condition by strand displacement activity of Bst DNA polymerase and visual read-outs (27-30). In fact, the efficiency of LAMP in diagnosis of pulmonary TB is evident from wide ranges of studies (31-35). Additionally, LAMP has been successfully deployed for diagnosis of other forms of TB such as tuberculous meningitis (36,37), osteoarticular TB (38), and tubercular lymphadenitis (39). Although a few studies have evaluated the diagnostic validity of LAMP by meta-analysis for diagnosis of MTB (40), pulmonary TB (41), and extrapulmonary TB (42), an updated meta-analysis covering all forms of mycobacteria was still missing. Hence, the aim of the present study was to systematically review and perform the meta-analysis to assess the diagnostic accuracy of the LAMP assay for detection of all forms of mycobacteria.
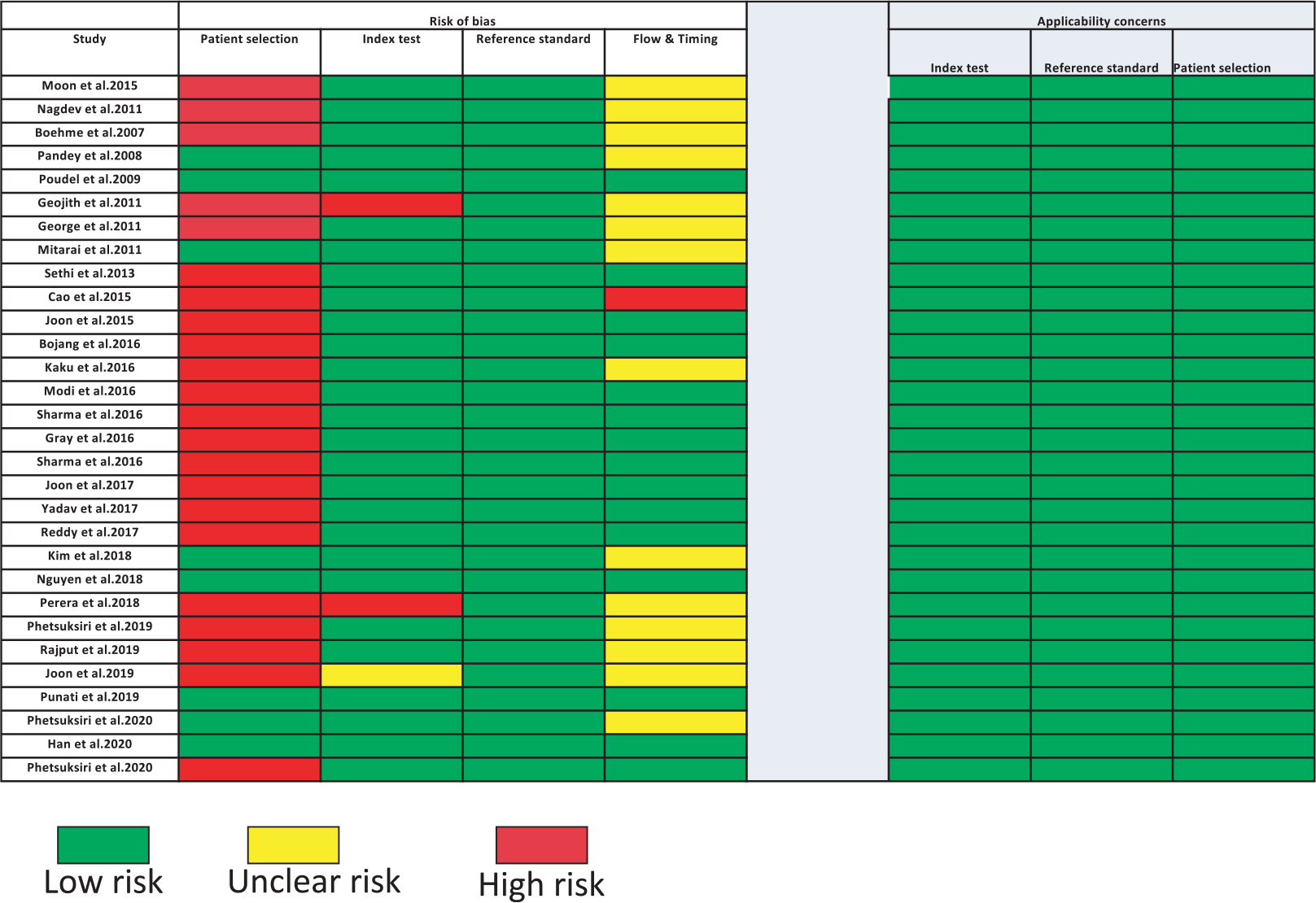
Fig. 8 - Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) summary of items for risk of bias and applicability in included studies (n = 30). Green color depicts the low risk of biasedness, yellow color depicts the unclear risk of biasedness, and red color depicts the high risk of biasedness.
This meta-analysis revealed that most of the studies were conducted in high TB burden countries such as India, Thailand, and Japan (Fig. 2). We observed that for the detection of mycobacteria sputum could be considered as the most chosen sample (Fig. 3). When considering the target genes, we found a variety of genes that were used in the included studies. However, IS6110 ranked first among all evaluated genes in the included studies (Fig. 4). This occurrence could be due to the presence of multiple copies of IS6110 present in the MTB genome (43). However, other target genes such as 16s rRNA and gyrB were also prominent. Next, we considered the detection method that was used for assessing the LAMP results. Most of the studies used fluorescence-based methods followed by colorimetry, gel electrophoresis, and turbidity, with no justification of their choices (Fig. 5). The prominence of fluorescence methods could be due to their increased sensitivity for the detection. Exceptionally, only one study mentioned lateral flow-based detection method despite market applicability.
Forest plot was used to calculate the sensitivity and specificity. The pooled sensitivity values of meta-analysis ranged between 0.26 and 1.0 (Fig. 6) and forest plot and SROC curve revealed a pooled specificity value between 0.67 and 1.0 (Fig. 7) with 95% CI. The accuracy and precision were calculated for the included studies and for 16 studies we found that the accuracy rate was higher than their corresponding precision rates and vice versa for 14 articles upon intra-comparison of accuracy with precision (Tab. II).
The current study also exhibited few limitations. Firstly, we observed high risk of patient selection bias or index test bias in almost two-thirds of the eligible studies (Fig. 8). Therefore, the use of unbiased patient cohorts and double-blinded index test may be recommended for future studies. Secondly, few studies showed the highest performance with 100% sensitivity and specificity, respectively, hence displaying the lowest QUADAS risk and concerns in all the domains. Furthermore, lack of subgroup analysis and the use of solely peer-reviewed English language articles were also additional limitations. Hence, although the current meta-analysis should be interpreted with caution, however, we believe that it will not impact the robustness of the analysis leading to further improved studies and reviews. Particularly considering the growing significance of LAMP-based detection for TB comparable to other methods, such studies may be encouraged (43-45).
Conclusion
Despite suffering from few disadvantages, like false positivity due to heavy reliance on indirect detection methods such as turbidity and nonspecific dyes and not providing any additional benefits like information on mutations, drug resistance etc., the LAMP technique could be a promising molecular test to enhance case detection before conventional time-consuming culture. Its simplicity, less turnaround time, and cost-effectiveness are major attractions for clinical laboratories. Also, it will be unjust to rely on single point-of-care test for TB successfully in various kinds of populations and resource availability. Although the unit cost is higher than smear microscopy and culture-based methods, it is likely to offer good value for money relative to conventional methods. In a nutshell, the present study endorses the use of LAMP assay as a promising alternative for detection of mycobacteria, particularly in regions which are financially compromised, where drug-resistant strains are not prevalent and PCR-based tests cannot be done so frequently. The faster diagnosis through LAMP could provide an alternative solution for failed medications to current therapeutics due to delayed diagnosis and subsequent development of drug resistance, thereby providing an opportunity to employ this new information in improving treatment strategies. However, the LAMP assay still must be improved to turn to a strong and competitive alternative to other molecular diagnostic methods.
Acknowledgments
We are grateful to Akansha Bhatt and Reva Gautam for their assistance in reviewing the literature and to Muriel Billamboz for assistance in English language editing.
Author contributions
G.S.B.: search, data extraction, validation. G.S.B., S.J.: data analysis. Z.F. and S.H.: supervision. G.S.B. and S.H.: writing, original draft. Z.F. and S.H. contributed to the conception and design of the study and review and editing of the manuscript.
Disclosures
Conflict of interest: The authors declare no conflicts of interest, financial or otherwise.
Financial support: Financial assistance to S.H. from the Indian Council of Medical Research (ICMR), New Delhi (5/8/5/9/ITRC/Diag/2022/ECD-1), is sincerely acknowledged.
References
- 1. Norbis L, Alagna R, Tortoli E, Codecasa LR, Migliori GB, Cirillo DM. Challenges and perspectives in the diagnosis of extrapulmonary tuberculosis. Expert Rev Anti Infect Ther. 2014;12(5):633-647. CrossRef PubMed
- 2. Pai M, Nicol MP, Boehme CC. Tuberculosis diagnostics: state of the art and future directions. Microbiol Spectr. 2016;4(5):1-15. CrossRef PubMed
- 3. World Health Organization. Global Tuberculosis Report 2015.WHO 2015.
- 4. American Thoracic Society; Centers for Disease Control and Prevention; Infectious Diseases Society of America. American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America: controlling tuberculosis in the United States. Am J Respir Crit Care Med. 2005;172(9):1169-1227. CrossRef PubMed
- 5. Park KS, Kim JY, Lee JW, et al. Comparison of the Xpert MTB/RIF and Cobas TaqMan MTB assays for detection of Mycobacterium tuberculosis in respiratory specimens. J Clin Microbiol. 2013;51(10):3225-3227. CrossRef PubMed
- 6. Notomi T, Okayama H, Masubuchi H, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63. CrossRef PubMed
- 7. Bhatt A, Fatima Z, Ruwali M, et al. CLEVER assay: a visual and rapid RNA extraction-free detection of SARS-CoV-2 based on CRISPR-Cas integrated RT-LAMP technology. J Appl Microbiol. 2022;133(2):410-421. CrossRef PubMed
- 8. Huang WE, Lim B, Hsu CC, et al. RT-LAMP for rapid diagnosis of coronavirus SARS-CoV-2. Microb Biotechnol. 2020;13(4):950-961. CrossRef PubMed
- 9. Broughton JP, Deng X, Yu G, et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38(7):870-874. CrossRef PubMed
- 10. World Health Organization. The use of loop-mediated isothermal amplification (TBLAMP) for the diagnosis of pulmonary tuberculosis: policy guidance. World Health Organization 2016;1-52.
- 11. Moher D, Liberati A, Tetzlaff J, Altman DG; PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. J Clin Epidemiol. 2009;62(10):1006-1012. CrossRef PubMed
- 12. Morillas AV, Gooch J, Frascione N. Feasibility of a handheld near infrared device for the qualitative analysis of bloodstains. Talanta. 2018;184:1-6. CrossRef PubMed
- 13. Gregório I, Zapata F, Torre M, García-Ruiz C. Statistical approach for ATR-FTIR screening of semen in sexual evidence. Talanta. 2017;174:853-857. CrossRef PubMed
- 14. Subali AD, Wiyono L. Reverse transcriptase loop mediated isothermal amplification (RT-LAMP) for COVID-19 diagnosis: a systematic review and meta-analysis. Pathog Glob Health. 2021;115(5):281-291. CrossRef PubMed
- 15. Subsoontorn P, Lohitnavy M, Kongkaew C. The diagnostic accuracy of isothermal nucleic acid point-of-care tests for human coronaviruses: a systematic review and meta-analysis. Sci Rep. 2020;10(1):22349. CrossRef PubMed
- 16. Phetsuksiri B, Klayut W, Rudeeaneksin J, et al. The performance of an in-house loop-mediated isothermal amplification for the rapid detection of Mycobacterium tuberculosis in sputum samples in comparison with Xpert MTB/RIF, microscopy and culture. Rev Inst Med Trop São Paulo. 2020;62:e36. CrossRef PubMed
- 17. Phetsuksiri B, Rudeeaneksin J, Srisungngam S, et al. Comparison of loop-mediated isothermal amplification, microscopy, culture, and PCR for diagnosis of pulmonary tuberculosis. Jpn J Infect Dis. 2020;73(4):272-277. CrossRef PubMed
- 18. Punati RD, Mallepaddi PC, Poonati R, et al. Development and evaluation of LAMP-coupled lateral flow device for the detection of MAP in livestock at point of care resource-limited areas. Braz J Microbiol. 2019;50(4):1105-1114. CrossRef PubMed
- 19. Geojith G, Dhanasekaran S, Chandran SP, Kenneth J. Efficacy of loop mediated isothermal amplification (LAMP) assay for the laboratory identification of Mycobacterium tuberculosis isolates in a resource limited setting. J Microbiol Methods. 2011;84(1):71-73. CrossRef PubMed
- 20. Nguyen VAT, Nguyen HV, Dinh TV, et al. Evaluation of Loopamp™ MTBC detection kit for diagnosis of pulmonary tuberculosis at a peripheral laboratory in a high burden setting. Diagn Microbiol Infect Dis. 2018;90(3):190-195. CrossRef PubMed
- 21. Han M, Xiao H, Yan L. Diagnostic performance of nucleic acid tests in tuberculous pleurisy. BMC Infect Dis. 2020;20(1):242. CrossRef PubMed
- 22. Perera SU, Navaratne V, Nagahawatte A, et al. Validating the loop mediated isothermal amplification (LAMP) technique to detect tuberculosis in a Sri Lankan laboratory setting. Ceylon Med J. 2018 31;63(1):40-42. CrossRef PubMed
- 23. Joon D, Nimesh M, Gupta S, Kumar C, Varma-Basil M, Saluja D. Development and evaluation of rapid and specific sdaA LAMP-LFD assay with Xpert MTB/RIF assay for diagnosis of tuberculosis. J Microbiol Methods. 2019;159:161-166. CrossRef PubMed
- 24. Cao D, Hu L, Lin M, et al. Real-time fluorescence loop-mediated isothermal amplification (LAMP) for rapid and reliable diagnosis of pulmonary tuberculosis. J Microbiol Methods. 2015;109:74-78. CrossRef PubMed
- 25. Bojang AL, Mendy FS, Tientcheu LD, et al. Comparison of TB-LAMP, GeneXpert MTB/RIF and culture for diagnosis of pulmonary tuberculosis in The Gambia. J Infect. 2016;72(3):332-337. CrossRef PubMed
- 26. Joon D, Nimesh M, Saluja D. Loop-mediated isothermal amplification as alternative to PCR for the diagnosis of extra-pulmonary tuberculosis. Int J Tuberc Lung Dis. 2015;19(8):986-991. CrossRef PubMed
- 27. Gray CM, Katamba A, Narang P, et al. Feasibility and operational performance of tuberculosis detection by loop-mediated isothermal amplification platform in decentralized settings: results from a multicenter study. J Clin Microbiol. 2016;54(8):1984-1991. CrossRef PubMed
- 28. Kaku T, Minamoto F, D’Meza R, et al. Accuracy of LAMP-TB method for diagnosing tuberculosis in Haiti. Jpn J Infect Dis. 2016;69(6):488-492. CrossRef PubMed
- 29. Kim CK, Cho EA, Shin DM, Choi SW, Shin SY. Comparative evaluation of the loop-mediated isothermal amplification assay for detecting pulmonary tuberculosis. Ann Lab Med. 2018;38(2):119-124. CrossRef PubMed
- 30. Mitarai S, Okumura M, Toyota E, et al. Evaluation of a simple loop-mediated isothermal amplification test kit for the diagnosis of tuberculosis. Int J Tuberc Lung Dis. 2011;15(9):1211-1217, i. CrossRef PubMed
- 31. Moon SH, Kim EJ, Tomono J, et al. Detection of Mycobacterium tuberculosis complex in sputum specimens using a loop-mediated isothermal amplification assay in Korea. J Med Microbiol. 2015;64(11):1335-1340. CrossRef PubMed
- 32. Pandey BD, Poudel A, Yoda T, et al. Development of an in-house loop-mediated isothermal amplification (LAMP) assay for detection of Mycobacterium tuberculosis and evaluation in sputum samples of Nepalese patients. J Med Microbiol. 2008;57(Pt 4):439-443. CrossRef PubMed
- 33. Phetsuksiri B, Rudeeaneksin J, Srisungngam S, et al. Loop-mediated isothermal amplification for rapid identification of Mycobacterium tuberculosis in comparison with immunochromatographic SD Bioline MPT64 Rapid® in a high burden setting. Jpn J Infect Dis. 2019;72(2):112-114. CrossRef PubMed
- 34. Sethi S, Singh S, Dhatwalia SK, et al. Evaluation of in-house loop-mediated isothermal amplification (LAMP) assay for rapid diagnosis of M. tuberculosis in pulmonary specimens. J Clin Lab Anal. 2013;27(4):272-276. CrossRef PubMed
- 35. Yadav R, Sharma N, Khaneja R, et al. Evaluation of the TB-LAMP assay for the rapid diagnosis of pulmonary tuberculosis in Northern India. Int J Tuberc Lung Dis. 2017;21(10):1150-1153. CrossRef PubMed
- 36. Modi M, Sharma K, Sharma M, et al. Multitargeted loop-mediated isothermal amplification for rapid diagnosis of tuberculous meningitis. Int J Tuberc Lung Dis. 2016;20(5):625-630. CrossRef PubMed
- 37. Nagdev KJ, Kashyap RS, Parida MM, et al. Loop-mediated isothermal amplification for rapid and reliable diagnosis of tuberculous meningitis. J Clin Microbiol. 2011;49(5):1861-1865. CrossRef PubMed
- 38. Sharma K, Sharma M, Batra N, Sharma A, Dhillon MS. Diagnostic potential of multi-targeted LAMP (loop-mediated isothermal amplification) for osteoarticular tuberculosis. J Orthop Res. 2017;35(2):361-365. CrossRef PubMed
- 39. Sharma M, Sharma K, Sharma A, Gupta N, Rajwanshi A. Loop-mediated isothermal amplification (LAMP) assay for speedy diagnosis of tubercular lymphadenitis: the multi-targeted 60-minute approach. Tuberculosis (Edinb). 2016;100:114-117. CrossRef PubMed
- 40. Nagai K, Horita N, Yamamoto M, et al. Diagnostic test accuracy of loop-mediated isothermal amplification assay for Mycobacterium tuberculosis: systematic review and meta-analysis. Sci Rep. 2016;6(1):39090. CrossRef PubMed
- 41. Shete PB, Farr K, Strnad L, Gray CM, Cattamanchi A. Diagnostic accuracy of TB-LAMP for pulmonary tuberculosis: a systematic review and meta-analysis. BMC Infect Dis. 2019;19(1):268. CrossRef PubMed
- 42. Yu G, Shen Y, Zhong F, Ye B, Yang J, Chen G. Diagnostic accuracy of the loop-mediated isothermal amplification assay for extrapulmonary tuberculosis: a meta-analysis. PLoS One. 2018;13(6):e0199290. CrossRef PubMed
- 43. Das S, Mangold KA, Shah NS, Peterson LR, Thomson RB Jr, Kaul KL. Performance and utilization of a laboratory-developed nucleic acid amplification test (NAAT) for the diagnosis of pulmonary and extrapulmonary tuberculosis in a low-prevalence area. Am J Clin Pathol. 2020;154(1):115-123. CrossRef PubMed
- 44. Tayal D, Sethi P, Jain P. Point-of-care test for tuberculosis – a boon in diagnosis. Monaldi Arch Chest Dis. 2023. CrossRef PubMed
- 45. Ludi Z, Sule AA, Samy RP, et al. Diagnosis and biomarkers for ocular tuberculosis: from the present into the future. Theranostics. 2023;13(7):2088-2113. CrossRef PubMed