 |
Drug Target Insights 2022; 16: 88-96 ISSN 1177-3928 | DOI: 10.33393/dti.2022.2522 REVIEW |
|
Current molecular approach for diagnosis of MRSA: a meta-narrative review
ABSTRACT
Introduction: Detection and diagnosis of methicillin-resistant Staphylococcus aureus (MRSA) are important in ensuring a correct and effective treatment, further reducing its spread. A wide range of molecular approaches has been used for the diagnosis of antimicrobial resistance (AMR) in MRSA. This review aims to study and appraise widely used molecular diagnostic methods for detecting MRSA.
Methods: This meta-narrative review was performed by searching PubMed using the following search terms: (molecular diagnosis) AND (antimicrobial resistance) AND (methicillin-resistant Staphylococcus aureus). Studies using molecular diagnostic techniques for the detection of MRSA were included, while non-English language, duplicates and non-article studies were excluded. After reviewing the libraries and a further manual search, 20 studies were included in this article. RAMESES publication standard for narrative reviews was used for this synthesis.
Results: A total of 20 full papers were reviewed and appraised in this synthesis, consisting of PCR technique (n = 7), deoxyribonucleic acid (DNA) Microarray (n = 1), DNA sequencing (n = 2), Xpert MRSA/SA BC assay (n = 2), matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) (n = 2), MLST (n = 4), SCCmec typing (n = 1) and GENECUBE (n = 1).
Discussion: Different diagnostic methods used to diagnose MRSA have been studied in this review. This study concludes that PCR has been extensively used due to its higher sensitivity and cost-effectiveness in the past five years
Keywords: Antimicrobial resistance, Molecular diagnosis, MRSA
Received: November 17, 2022
Accepted: December 31, 2022
Published online: December 31, 2022
Drug Target Insights - ISSN 1177-3928 - www.aboutscience.eu/dti
© 2022 The Authors. This article is published by AboutScience and licensed under Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0).
Commercial use is not permitted and is subject to Publisher’s permissions. Full information is available at www.aboutscience.eu
Introduction
Antimicrobial resistance (AMR) is defined as changes in bacteria that result in the drug being used for its treatment becoming inefficacious (1). Staphylococcus aureus is an opportunistic pathogen with a tremendous capacity to adapt to human hosts and healthcare environments, causing detrimental effects to healthcare-associated infections such as bloodstream infections (2). AMR is reported as the world’s biggest 21st-century health threat, and the World Health Organization (WHO) is calling for immediate action. As AMR spreads, common infections are becoming incurable. Reports state that over 700,000 die yearly due to drug-resistant illnesses; by 2050, the number is predicted to rise to 10 million (3).
A major issue pertaining to AMR is the excessive and injudicious use of antibiotics that have led to widespread resistant bacteria and dissemination of their antimicrobial resistant genes (ARGs) (4). It is concerning that the AMR rates are predicted to increase if measures are not taken. One way to overcome this is through early detection, which enables effective management, allowing efficient identification and detection of microbes such that the patient can be treated with the appropriate drug in time.
Over the years, great leaps have been made in the diagnosis of AMR and diagnostic tests are reported to be an essential tool in early diagnosis, hence it is a robust strategy against AMR (4).
To enhance existing approaches, this review aims to summarize new and current molecular techniques and technologies used to identify AMR using a systematic meta-narrative approach, with a focus on the key benefits and drawbacks. Furthermore, a critical overview of recently developed molecular approaches and an informed assessment of future direction will also be discussed.
Methodology
Study design and inclusion criteria
This systematic review was carried out in a meta-narrative framework. This study qualitatively appraised different molecular methods used in the recent 5 years for the diagnosis of methicillin-resistant Staphylococcus aureus (MRSA). This study protocol was created according to the RAMESES (Realist And Meta-narrative Evidence Syntheses: Evolving Standards) meta-narrative review publication guidelines (5). Articles that satisfied the following requirements were considered for the review: (i) original articles written in English that were published between January 2017 and May 2022, (ii) cross-sectional or cohort studies that assessed the technical performance of molecular methods (sensitivity, specificity, accuracy or concordance) for diagnosing MRSA. Articles were excluded if they were: (i) case reports; (ii) review articles, commentary articles, and short communications.
Search strategies
Articles were searched using PubMed. Search keywords were (((((molecular diagnosis) AND (antimicrobial resistance)) NOT (review [publication type)) NOT (systematic review [publication type)) NOT (meta-analysis [publication type)) AND (methicillin-resistant staphylococcus aureus).
Selection and appraisal of articles
Two independent reviewers (Lee and Sim) screened the titles and abstracts. Articles with abstracts indicating the use of a molecular approach to diagnose MRSA were read in full. A final consensus was discussed between the two reviewers, and disagreements were resolved with discussion from the third reviewer (SM). EndNote Version 20 was used for article duplicate removal and archives. All the studies reviewed and appraised in this synthesis are summarized in Table I.
| No | Author | Year | Country | Condition/patients | Sample | Study design | Molecular diagnosis methods | Reference |
|---|---|---|---|---|---|---|---|---|
| 1 | Moutaouakkil et al | 2022 | China | Children diagnosed with Staphylococcus aureus OAI | Blood cultures, articular fluids, synovial tissues and/or bone fragments | Prospective study | Multiplex polymerase chain reaction | (6) |
| 2 | Jin et al | 2022 | China | 1,952 MSSA strains isolated from blood across 17 provinces | MSSA-PENS isolated from invasive BSIs | Retrospective study | Whole-genome sequencing | (2) |
| 3 | Senok et al | 2021 | United Arab Emirates | 135 patients with a clinical diagnosis of severe skin and soft-tissue infections | S. aureus isolates associated with SSTI were tested for PVL detection | n/a | DNA microarray assays | (7) |
| 4 | Reddy and Whitelaw | 2021 | South Africa | 231 samples | 2,822 patients with positive blood cultures exclusively showing GPCC on Gram stain were included | Prospective study | Xpert MRSA/SA BC assay | (8) |
| 5 | Choi et al | 2021 | South Korea | 26 children aged <15 years diagnosed with SSSS | Involved area of the skin, the presence of Nikolsky’s sign, and the status of desquamation | n/a | PCR | (9) |
| 6 | Anafo et al | 2021 | Ghana | 300 diabetes patients and 106 non-diabetic individuals | Anterior nasal swabs | Cross-sectional | PCR | (10) |
| 7 | Verdú-Expósito et al | 2020 | Ethiopia | 80 S. aureus strains isolated from human patients with SSTIs | Human samples | n/a | MALDI-TOF and PCR | (11) |
| 8 | Tang et al | 2020 | China | MRSE strains from the dental plaque of a normal, healthy human population | Dental plaque specimens | n/a | PCR | (12) |
| 9 | Khawaja et al | 2020 | Pakistan | 105 samples | Human samples | Descriptive cross-sectional study | PCR | (13) |
| 10 | Jin et al | 2020 | China | 65-Year-old healthy man with a history of leprosy | Isolate was obtained from the patient’s blood, and identified as an ST9-MRSA strain | n/a | Whole-genome sequencing | (14) |
| 11 | Geng et al | 2020 | China | 536 neonates | Nasal swabs | Prospective surveillance study | Staphylococcal chromosomal cassette (and) type, spa type, MLST | (15) |
| 12 | Crandall et al | 2020 | USA | 357 children with invasive S. aureus infections | Pleural fluid and/or blood | Prospective study | PCR, MLST, SCCmec typing | (16) |
| 13 | Bouza et al | 2020 | Spain | 155 adult inpatients diagnosed with skin and soft-tissue infection | Microbiological samples | Prospective study | Gram stain plus GeneXpert® MSSA/MRSA SSTI | (17) |
| 14 | Yang et al | 2019 | China | 269 nonduplicate S. aureus clinical isolates were isolated from children | Steril specimens and non-STERIL specimen using VITEK MS system | n/a | MALDI-TOF | (18) |
| 15 | Mutonga et al | 2019 | Kenya | 83 adult patients diagnosed with diabetic foot ulcers | Wound swab cultures | Cross-sectional study | Real-time PCR | (19) |
| 16 | Latour et al | 2019 | Belgium | 1,447 residents from nursing homes | Pooled sampling of nose, throat and perineum | Cross-sectional prevalence survey | Triplex PCR and MLST | (20) |
| 17 | Hida et al | 2019 | Japan | 263 patients suspected of having staphylococcal bacteremia | Fresh and frozen blood culture samples | n/a | GENECUBE mecA | (21) |
| 18 | Luo et al | 2018 | China | 275 isolates of S. aureus, including 148 isolates from patients, 127 from ready-to-eat food samples | Secretions, blood, phlegm, cerebrospinal fluid, transudation, urine, fresh meat, meat product, cereal products, fruits and vegetables | n/a | PCR, multiplex PCR | (22) |
| 19 | Lin et al | 2018 | Taiwan | 106 hemodialysis patients diagnosed with MRSA | Blood cultures | Retrospective study | PCR and MLST | (23) |
| 20 | Yang et al | 2017 | China | 104 children diagnosed with MRSA | Sputum, bronchioalveolar lavage fluid, skin and soft tissues, pus, secretions, secretions of omphalitis, blood, joint effusion, pleural effusion | n/a | MLST | (24) |
BSI = bloodstream infection; DNA = deoxyribonucleic acid; MALDI-TOF = matrix-assisted laser desorption/ionization-time of flight; MLST = multilocus sequence typing; MRSA = methicillin-resistant Staphylococcus aureus; MRSE = methicillin-resistant Staphylococcus epidermidis; MSSA = methicillin-sensitive Staphylococcus aureus; n/a = not available; PCR = polymerase chain reaction; SCCmec = staphylococcal cassette chromosome mec; spa = staphylococcal protein A.
GPCC = Gram positive cocci in clusters; MSSA-PENS = methicillin-sensitive S. aureus – penicillin-susceptible; OAI = osteoarticular infections; SSSS = Staphylococcal scalded skin syndrome ; SSTI = skin and soft tissue infections; PVL = Panton Valentine leukocidin
Results
The dataset includes 20 different authors from Asia (n = 13), Africa (n = 5), Europe (n = 1) and America (n = 1). A total of 20 studies were included in this synthesis: seven studies employed polymerase chain reaction (PCR) for diagnosing MRSA (6,9,10,12,13,19,22), one study employed deoxyribonucleic acid (DNA) Microarray (7), two studies used DNA sequencing (2,14), Xpert MRSA/SA BC assay (n = 2) (8,17), matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF; n = 2) (11,18), multilocus sequence typing (MLST; n = 4) (15,20,23,24), GENECUBE (n = 1) (21) and staphylococcal cassette chromosome mec (SCCmec) typing (n = 1) (16). Figure 1 is the diagrammatic flow of the study selection and list of techniques appraised in this review.
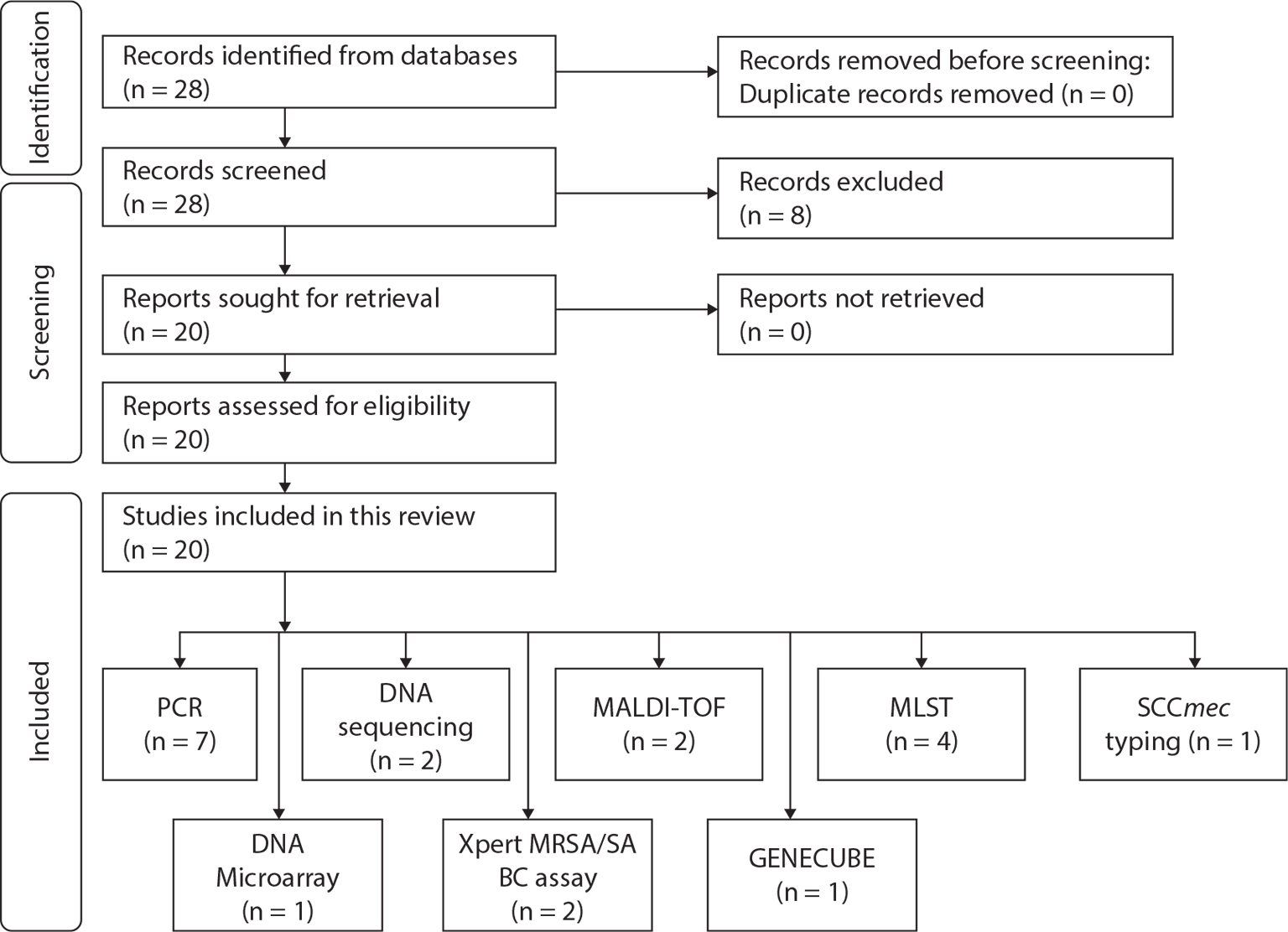
Fig. 1 - Diagrammatic flow of the study selection and list of techniques appraised in this review.
Recent molecular methods for diagnosis of MRSA
Polymerase chain reaction
PCR approaches have been commonly used for the effective diagnosis of MRSA, and the rapid emergence of MRSA has led to a series of PCR approaches that have been developed for the identification of MRSA (25). PCR approach identifies S. aureus based on a single-base-pair mismatch in the staphylococcal 16S ribosomal RNA gene sequence (26). Recent researchers have also cited the use of the PCR approach for mecA gene detection as the gold standard method for the detection and identification of the prevalence of MRSA (27,28). In this synthesis, a total of seven studies have employed PCR for the detection and diagnosis of MRSA. A study conducted by Moutaouakkil and colleagues among patients suspected of S. aureus hospitalized in pediatric orthopedic clinic reported the detection of mecA using PCR (6). This study also utilized different biological samples such as blood cultures, articular fluids, synovial tissues and bone fragments for the detection of MRSA. Another study showed that the fluorescence signal of real-time (RT)-PCR could display the quantity of products formed and increases exponentially, enabling a user-friendly diagnostic (29). Furthermore, Mutonga and colleagues (2019) have demonstrated that the sensitivity of RT-PCR for MRSA is 100% (19).
Multiplex PCR amplifies multiple DNA sequences simultaneously, which gives an advantage over conventional PCR (30). The detection of target sequences, such as the nuc and coaA or elements necessary for methicillin resistance, such as femA, or femB, has provided the basis for PCR identification of S. aureus. It uses two pairs of primers specific to the staphylococcal nuc and mecA for PCR amplification of a 280-bp nuc-based fragment and a 533-bp mecA-based fragment (31). Tsai and colleagues (2019) reported mecA gene (mecA-F and mecA-R) is amplified and can be used to diagnose MRSA (32). Chikkala and colleagues showed that it exhibits 97% of specificity and 90% sensitivity (33).
DNA sequencing
DNA sequencing allows the detection of single-nucleotide polymorphisms (SNPs) and known resistance-associated genes and their variations (34). The availability of bacterial genomes in public databases facilitates the use of whole-genome sequencing for MRSA detection. It enables high-resolution characterization of antibiotic resistance (35). Whole-genome sequencing has a definite edge over conventional Sanger sequencing because it may produce millions of reads that are roughly 35 to 700 bp in length (36). There is growing evidence on the effectiveness of bacterial whole-genome sequencing in controlling outbreaks. Whole-genome analysis, such as DNA microarray, simultaneously identifies relative concentration of different nucleic acid sequence (37). It allows a bulk number of nucleic acid sequences in a mixture to be tested and analyzed. The study by Jin and colleagues (2,14) used StaphyType DNA microarray (Abbott [Alere Technologies GmbH], Jena, Germany) and the INTER-ARRAY Genotyping Kit S. aureus (Inter-Array GmbH, Bad Langensalza, Germany) for the detection of MRSA. The study by Senok and colleagues (2021) also reported that DNA microarray exhibited 100% specificity and sensitivity (7). In a study done by Ma and fellow colleagues, Illumina’s Nextera DNA library preparation kit was used to create whole-genome sequencing libraries, which were then sequenced on an Illumina MiSeq using the 500 cycle V2 protocol (38).
Xpert MRSA/SA BC assay
Xpert MRSA/SA Blood Culture is an in vitro diagnostic test for S. aureus and MRSA. The targeted DNA is amplified using automated RT-PCR and Fluorogenic target-specific hybridization, providing real-time detection of specific genes of MRSA and S. aureus. A study by Buchan and colleagues (39) reported the use of blood cultures for the detection of Staphylococcus protein A (spa) sequences, gene that encodes for methicillin resistance (mecA) and SCCmec. A study by Reddy and colleagues has shown the performance of the Xpert MRSA/SA BC assay to be 100% in specificity and sensitivity. It shows a failure rate for an interpretable result of just 1.7% (8). However, it is notable that the microbiological sampling should be of high quality to ensure rapid and accurate results, despite the significance of Xpert MRSA system.
MALDI-TOF
MALDI-TOF mass spectrometry (MS) has become a widely used technique for the rapid and accurate identification of bacteria (40). Despite the efficiency and sensitivity of MALDI-TOF, this method’s limitation is that new isolates can only be detected if the spectral database contains peptide mass fingerprints (PMFs) of the type strains of specific genera/species/subspecies/strains. This method identifies microbes by comparing the PMF of unknown organisms with the PMFs deposited in the database or matching the masses of biomarkers with the proteome database. A recent study by Tang and colleagues (41) reported that MALDI-TOF MS on intact bacteria combined with a refined analysis framework allows accurate classification of methicillin-sensitive Staphylococcus aureus (MSSA) and MRSA. Esener and colleagues showed that MALDI-TOF has a sensitivity of 99.93% ± 0.25%, specificity of 95.04% ± 3.83%, and accuracy = 97.54% ± 1.91% (42). MALDI-TOF is low in cost, and analysis can be conducted within a short time, allowing rapid microbial resistance to be detected. Latour and colleagues employed MALDI BioTyper database for bacterial identification of suspected colonies (20). A study by Chen and colleagues has shown that MLST has been used for the past decades for MRSA epidemiological typing (43). However, it is only based on the sequences of seven house-keeping genes’ internal fragments to identify individual isolate lineages.
MLST
MLST is a technique that distinguishes between isolates of bacteria species by utilizing sequences of internal fragment house-keeping genes (44). The strands are sequenced on both side by using an automated DNA sequencer. Different sequences of house-keeping genes found in bacterial species are characterized as distinct alleles. In contrast, seven loci alleles address each isolate’s allelic profile or sequence type. Hence, species isolates are unambiguously characterized by a series of seven integers which label the alleles at the seven house-keeping genes. The seven house-keeping genes used in MLST for S. aureus are the Carbamate kinase (arcC), Shikimate dehydrogenase (aroE), glycerol kinase (glpF), Guanylate kinase (gmk), Phosphate acetyltransferase (pta), Triosephosphate isomerase (tpi), acetyl coenzyme A acetyltransferase (18,24,45).
SPA typing
Spa is an important gene virulence factor that allows S. aureus to avoid host immune responses (46). It codes for protein A, which is found in the cell wall of S. aureus (47). SPA genes were replicated using PCR followed by DNA sequencing (48). This method identifies the polymorphic X region of the protein A gene (spa). Based Upon Repeat Pattern (BURP) algorithm was used, and spa types with more than five repeats were clustered into different groups, with the calculated cost between group members being less than or equal to 6 (49). Spa typing is evidently reproducible and provides interchangeable information. However, a disadvantage of this method is that it requires additional targets such as SCCmec, lineage-specific virulence or resistance genes or alternative polymorphic regions of the S. aureus chromosome. Studies included in this synthesis employed Ridom Staph Database and SPA typer tool (http://spatyper.fortinbras.us/) (24,50,51). Reports cited that spa type of t437 was more prevalent in MRSA (24). A study by Luo and colleagues showed that the most prominent spa type was t030, reported to be 15.64% (43/275) (22).
GENECUBE assays
GENECUBE (TOYOBO Co., Ltd., Osaka, Japan) is a fully automated genetic analyzer that uses PCR to amplify a target gene (21). This tool can evaluate up to eight samples simultaneously. The target DNA is amplified, and fluorescently labeled oligonucleotides are used to hybridize targets based on fluorescence intensity changes (52). Data are automatically obtained on the GENECUBE monitor after completion of the assay. The advantage of this assay is that it is time efficient and easy to prepare. GENECUBE tests are anticipated to be clinically valuable for effectively identifying MRSA. Studies have reported the sensitivity and specificity of the GENECUBE to be 100% (33). The system is accurate, rapid (52 minutes), and reliable; however, it does not detect the mecC gene (21).
SCCmec typing
SCCmec is a diagnostic method that divides SCCmec elements into groups based on their structural variations (53). The mec complex, which comprises the mec gene, its regulatory genes, the mecI and mecR1 genes, and several insertion sequences, confers methicillin resistance (54,55). The specific SCCmec type is determined by combining the ccr gene complex and the mec gene class. SCCmec typing provides valuable information about the resistance of genes to methicillin and identifies the origin of strains. A recent study by Chongtrakool et al (56) typed SCCmec of methicillin-resistant S. aureus strains isolated in 11 Asian countries. Another study showed that 610 of 615 (99.2%) MRSA strains could be classified into four SCCmec elements: type 3A, 370 strains; type 2A, 207; type 2B, 32; type 1B, 1 strain. This study on pandemic MRSA clones in Asia reported the ST59-SCCmecIVa as the most prevalent MRSA clone (15). A study by Chen and colleagues that used the web-based SCCmecFinder reported that this technique is efficient for detecting MRSA (43). SCCmecFinder is a web-based tool for SCCmec typing using whole-genome sequences (https://cge.cbs.dtu.dk/services/SCCmecFinder/, accessed on January 11, 2023). The SCCmecFinder website uses read data for whole-genome sequencing or preassembled genome/contigs to determine homology to the complete cassette in prediction of SCCmec types, mec complex and J regions (57).
Discussion
This meta-narrative review reports the commonly used molecular methods for the detection of MRSA in the past 5 years. This review has also summarized the advantages and disadvantages of each technique included in this synthesis.
S. aureus is a common cause of community and hospital-acquired infection (58,59). The WHO has regarded it as one of the primary clinical concerns, due to the global recognition of MRSA as a public health issue and the antibiotic resistance pattern of MRSA (60). The primary issue with MRSA is the incidence of multidrug resistance, which remains high (61).
The mecA encodes penicillin-binding protein 2a (PBP2a), which is an enzyme responsible for crosslinking peptidoglycans in the bacterial cell wall (62). The low affinity of PBP2a for β-lactams leads to resistance to β-lactam antibiotics, including penicillins, cephalosporins (except ceftaroline and ceftobiprole) and carbapenems (63). Recent reports have reported growing resistance to clindamycin and levofloxacin, necessitating an effective treatment.
The virulence factor of S. aureus is multifactorial and depends on a variety of toxins, adhesion, immune evasion and other virulence characteristics (64). Evaluation of the virulence factor is an effective method of predicting how these bacteria would behave in the host, enabling prediction of the onset and progression of an infection. The first stage of staphylococcal infection is when the bacterial cells connect to the host’s tissues. The surface-exposed proteins, MSCRAMMs (microbial surface components recognizing adhesive matrix molecules), are made by S. aureus, which functions to attach to one or more host extracellular matrix (ECM) components, such as laminin, elastin, fibrinogen, fibronectin and collagen (65,66). The extracellular adherence protein (Eap) produced by S. aureus is a member of the SERAMs (secretable expanded repertoire adhesive molecules) family, binds to ECM glycoproteins, including fibronectin, fibrinogen, sialoprotein and collagens (67). This protein is involved in the internalization of bacteria and the adherence of S. aureus to fibroblasts. Proteases are crucial virulence factors for S. aureus and can cleave host proteins to enable MRSA cells to change from an adhesive to an invasive phenotype.
Early diagnostic and therapeutic intervention in patients with MRSA infection risk factors is essential (68). Treatment with empiric antibiotics against MRSA should not be delayed in the event that MRSA infection is diagnosed. Molecular diagnostic tests can robustly identify staphylococcal species in clinical samples, thus improving antimicrobial stewardship (69).
In this review, multiple molecular methods such as PCR, DNA sequencing, Xpert MRSA/SA BC array, MALDI-TOF, MLST, SPA typing and SCCmec typing, have been appraised. This review summarizes that PCR technique has been widely used for the diagnosis of MRSA within the last 5 years (2017-2022).
PCR technique is frequently and commonly used to detect S. aureus and it identifies a single-base-pair mismatch in the staphylococcal 16S ribosomal RNA gene sequence for detection (26). PCR assay is cost and labor effective and can be conducted within a short period of time (70,71). However, studies have reported that different target genes may impact the specificity and sensitivity of PCR for diagnosis. The nuc gene has a 100% success rate (25,72). Several PCR techniques such as multiplex PCR, RT-PCR and isothermal identification have been developed to identify MRSA as a result of its rapid emergence. The mecA and nuc genes are being used due to their 100% sensitivity and 97% specificity respectively with a shorter turnaround time of 48 hours (73,74).
The second commonly used molecular techniques are SCCmec typing and MLST, respectively. Over the years, the structures of novel SCCmec have been identified and verified by molecular cloning and traditional sequencing (75). In a study by Singh-Moodley and colleagues (76), SCCmec typing method was used to replace multiplex PCR and was employed to classify additional un-typeable SCCmec elements based on ccr and mec gene complex combinations. However, this technique has been deemed highly complex because the SCCmec region is variable and newer types are permanently being developed. Another possible reason for using SCCmec typing could be its potential as a benchmark for testing for the ccr gene and mecA gene compared to other methods.
MLST is well-established and assigns alleles at multiple house-keeping loci directly by DNA sequencing. Sequence type is obtained based on the alleles identified at each of the seven loci using the SA MLST database. MLST detection of MRSA is based on the sequencing of the seven house-keeping conserved genes in the bacterial chromosome (77). MLST is also widely used due to its straightforward procedure for characterizing isolates of bacterial species (78). Due to numerous alleles in each of the seven loci, it is unlikely that two isolates will have the same allelic profile. Instead, isolates with the same allelic profile can be identified as belonging to the exact clone. MLST has several advantages: (1) it uses sequence data to detect changes at the DNA level; (2) it is readily reproduced and does not require specialized reagents or training; (3) it does not require high-quality genomic DNA; and (4) the data generated are fully portable (79). The disadvantage of MLST is that it only uses seven genes, limiting its ability.
DNA microarray and Xpert MRSA/SA BC assay are the least used in the last 5 years. DNA microarray contains covalently immobilized probes specific for about 180 genes and 300 alleles of S. aureus (80). It allows simultaneous detection of the presence of numerous genomic loci. Studies have reported that DNA microarray may serve as an alternate molecular typing method, offering complementary characterization of the MRSA strains. However, this technique is labor and cost extensive and a single experiment could significantly increase the budget of the experiment. Subsequently, many probe designs are based on a sequence of relatively low specificity, sensitivity and accuracy (81).
Conclusion
This meta-narrative review has appraised and summarized molecular diagnostic methods frequently used to detect MRSA in the last 5 years (2017-2022), thus concluding that PCR technique is the most frequently used technique due to its high specificity, low cost and labor effectiveness.
Acknowledgment
The authors would like to acknowledge the Higher Colleges of Technology Interdisciplinary Research Grant (Interdisciplinary_212322).
Disclosure
Conflict of interest: All authors declare no conflict of interest.
References
- 1. Murray CJ, Ikuta KS, Sharara F, et al; Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629-655. CrossRef PubMed
- 2. Jin Y, Zhou W, Zhan Q, et al. Genomic epidemiology and characterisation of penicillin-sensitive Staphylococcus aureus isolates from invasive bloodstream infections in China: an increasing prevalence and higher diversity in genetic typing be revealed. Emerg Microbes Infect. 2022;11(1):326-336. CrossRef PubMed
- 3. Hanson C, Gabrysch S, Mbaruku G, Cox J, Mkumbo E, Manzi F, Schellenberg J, Ronsmans C. Access to maternal health services: geographical inequalities, United Republic of Tanzania. Bull World Health Organ. 2017 Dec 1;95(12):810-820. CrossRef Epub 2017 Oct 31. PubMed PMCID: PMC5710083.
- 4. Kaprou GD, Bergšpica I, Alexa EA, Alvarez-Ordóñez A, Prieto M. Rapid methods for antimicrobial resistance diagnostics. Antibiotics (Basel). 2021;10(2):209. CrossRef PubMed
- 5. Wong G, Greenhalgh T, Westhorp G, Buckingham J, Pawson R. RAMESES publication standards: meta-narrative reviews. J Adv Nurs. 2013;69(5):987-1004. CrossRef PubMed
- 6. Moutaouakkil K, Abdellaoui H, Arhoune B, et al. Paediatric osteoarticular infections caused by Staphylococcus aureus producing panton-valentine leucocidin in Morocco: risk factors and clinical features. Afr J Paediatr Surg. 2022;19(2):78-82. CrossRef PubMed
- 7. Senok A, Monecke S, Nassar R, et al. Lateral flow immunoassay for the detection of panton-valentine leukocidin in Staphylococcus aureus from skin and soft tissue infections in the United Arab Emirates. Front Cell Infect Microbiol. 2021;11:754523. CrossRef PubMed
- 8. Reddy K, Whitelaw A. Can the Xpert MRSA/SA BC assay be used as an antimicrobial stewardship tool? A prospective assay validation and descriptive impact assessment study in a South African setting. BMC Infect Dis. 2021;21(1):177. CrossRef PubMed
- 9. Choi JH, Lee H, Choi EH. Antimicrobial resistance and molecular analysis of Staphylococcus aureus in staphylococcal scalded skin syndrome among children in Korea. J Korean Med Sci. 2021;36(3):e22. CrossRef PubMed
- 10. Anafo RB, Atiase Y, Kotey FCN, et al. Methicillin-resistant Staphylococcus aureus (MRSA) nasal carriage among patients with diabetes at the Korle Bu Teaching Hospital. PLoS One. 2021;16(9):e0257004. CrossRef PubMed
- 11. Verdú-Expósito C, Romanyk J, Cuadros-González J, et al. Study of susceptibility to antibiotics and molecular characterization of high virulence Staphylococcus aureus strains isolated from a rural hospital in Ethiopia. PLoS One. 2020;15(3):e0230031. CrossRef PubMed
- 12. Tang B, Gong T, Cui Y, et al. Characteristics of oral methicillin-resistant Staphylococcus epidermidis isolated from dental plaque. Int J Oral Sci. 2020;12(1):15. CrossRef PubMed
- 13. Khawaja A, Arshad F, Khan I. Comparison of phenotypic methods with mecA gene based polymerase chain reaction for methicillin-resistant Staphylococcus aureus detection. J Pak Med Assoc. 2020;70(2):276-280. PubMed
- 14. Jin Y, Yu X, Chen Y, et al. Characterization of highly virulent community-associated methicillin-resistant Staphylococcus aureus ST9-SCCmec XII causing bloodstream infection in China. Emerg Microbes Infect. 2020;9(1):2526-2535. CrossRef PubMed
- 15. Geng W, Qi Y, Li W, et al. Epidemiology of Staphylococcus aureus in neonates on admission to a Chinese neonatal intensive care unit. PLoS One. 2020;15(2):e0211845. CrossRef PubMed
- 16. Crandall H, Kapusta A, Killpack J, et al. Clinical and molecular epidemiology of invasive Staphylococcus aureus infection in Utah children; continued dominance of MSSA over MRSA. PLoS One. 2020;15(9):e0238991. CrossRef PubMed
- 17. Bouza E, Onori R, Semiglia-Chong MA, Álvarez-Uría A, Alcalá L, Burillo A. Fast track SSTI management program based on a rapid molecular test (GeneXpert® MRSA/SA SSTI) and antimicrobial stewardship. J Microbiol Immunol Infect. 2020;53(2):328-335. CrossRef PubMed
- 18. Yang X, Dong F, Qian S, et al. Accessory gene regulator (agr) dysfunction was unusual in Staphylococcus aureus isolated from Chinese children. BMC Microbiol. 2019;19(1):95. CrossRef PubMed
- 19. Mutonga DM, Mureithi MW, Ngugi NN, Otieno FCF. Bacterial isolation and antibiotic susceptibility from diabetic foot ulcers in Kenya using microbiological tests and comparison with RT-PCR in detection of S. aureus and MRSA. BMC Res Notes. 2019;12(1):244. CrossRef PubMed
- 20. Latour K, Huang TD, Jans B, et al. Prevalence of multidrug-resistant organisms in nursing homes in Belgium in 2015. PLoS One. 2019;14(3):e0214327. CrossRef PubMed
- 21. Hida Y, Uemura K, Sugimoto H, et al. Evaluation of performance of the GENECUBE assay for rapid molecular identification of Staphylococcus aureus and methicillin resistance in positive blood culture medium. PLoS One. 2019;14(7):e0219819. CrossRef PubMed
- 22. Luo K, Shao F, Kamara KN, et al. Molecular characteristics of antimicrobial resistance and virulence determinants of Staphylococcus aureus isolates derived from clinical infection and food. J Clin Lab Anal. 2018;32(7):e22456. CrossRef PubMed
- 23. Lin S-Y, Tu HP, Chen TC, et al. Association of bacterial genotypes and epidemiological features with treatment failure in hemodialysis patients with methicillin-resistant Staphylococcus aureus bacteremia. PLoS One. 2018;13(6):e0198486. CrossRef PubMed
- 24. Yang X, Qian S, Yao K, et al. Multiresistant ST59-SCCmec IV-t437 clone with strong biofilm-forming capacity was identified predominantly in MRSA isolated from Chinese children. BMC Infect Dis. 2017;17(1):733. CrossRef PubMed
- 25. Liu Y, Zhang J, Ji Y. PCR-based approaches for the detection of clinical methicillin-resistant Staphylococcus aureus. Open Microbiol J. 2016;10(1):45-56. CrossRef PubMed
- 26. Saruta K, Hoshina S, Machida K. Genetic identification of Staphylococcus aureus by polymerase chain reaction using single-base-pair mismatch in 16S ribosomal RNA gene. Microbiol Immunol. 1995;39(11):839-844. CrossRef PubMed
- 27. Woods SE, Beiter E, Drake B, Engel A. The prevalence of asymptomatic methicillin-resistant Staphylococcus aureus in school-age children. East J Med. 2011;16(1):18-21.
- 28. Sahebnasagh R, Saderi H, Owlia P. The prevalence of resistance to methicillin in Staphylococcus aureus strains isolated from patients by PCR method for Detection of mecA and nuc genes. Iran J Public Health. 2014;43(1):84-92. PubMed
- 29. Kubista M, Andrade JM, Bengtsson M, et al. The real-time polymerase chain reaction. Mol Aspects Med. 2006;27(2-3):95-125. CrossRef PubMed
- 30. Markoulatos P, Siafakas N, Moncany M. Multiplex polymerase chain reaction: a practical approach. J Clin Lab Anal. 2002;16(1):47-51. CrossRef PubMed
- 31. Barski P, Piechowicz L, Galiński J, Kur J. Rapid assay for detection of methicillin-resistant Staphylococcus aureus using multiplex PCR. Mol Cell Probes. 1996;10(6):471-475. CrossRef PubMed
- 32. Tsai YH, Chen PH, Yu PA, Chen CL, Kuo LT, Huang KC. A multiplex PCR assay for detection of Vibrio vulnificus, Aeromonas hydrophila, methicillin-resistant Staphylococcus aureus, Streptococcus pyogenes, and Streptococcus agalactiae from the isolates of patients with necrotizing fasciitis. Int J Infect Dis. 2019;81:73-80. CrossRef PubMed
- 33. Chikkala R, Ch S, Divyakolu S, Ratnakar KS, Sritharan V. A simple sample processing protocol and multiplex PCR for direct detection of MRSA from uncultured clinical samples – a pilot study. Adv Infect Dis. 2019;9(1):25-38. CrossRef
- 34. Sanchini A. Recent developments in phenotypic and molecular diagnostic methods for antimicrobial resistance detection in Staphylococcus aureus: a narrative review. Diagnostics (Basel). 2022;12(1):208. CrossRef PubMed
- 35. Gautam SS, Kc R, Leong KW, Mac Aogáin M, O’Toole RF. A step-by-step beginner’s protocol for whole genome sequencing of human bacterial pathogens. J Biol Methods. 2019;6(1):e110. CrossRef PubMed
- 36. Lakhundi S, Zhang K. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev. 2018;31(4):e00020-18. CrossRef PubMed
- 37. Bumgarner R. Overview of DNA microarrays: types, applications, and their future. Curr Protoc Mol Biol. 2013 Jan;Chapter 22:Unit 22.1. CrossRef
- 38. Ma Z, Lasek-Nesselquist E, Lu J, et al. Characterization of genetic changes associated with daptomycin nonsusceptibility in Staphylococcus aureus. PLoS One. 2018;13(6):e0198366. CrossRef PubMed
- 39. Buchan BW, Allen S, Burnham CA, et al. Comparison of the next-generation Xpert MRSA/SA BC assay and the GeneOhm StaphSR assay to routine culture for identification of Staphylococcus aureus and methicillin-resistant S. aureus in positive-blood-culture broths. J Clin Microbiol. 2015;53(3):804-809. CrossRef PubMed
- 40. Feucherolles M, Poppert S, Utzinger J, Becker SL. MALDI-TOF mass spectrometry as a diagnostic tool in human and veterinary helminthology: a systematic review. Parasit Vectors. 2019;12(1):245. CrossRef PubMed
- 41. Tang W, Ranganathan N, Shahrezaei V, Larrouy-Maumus G. MALDI-TOF mass spectrometry on intact bacteria combined with a refined analysis framework allows accurate classification of MSSA and MRSA. PLoS One. 2019;14(6):e0218951. CrossRef PubMed
- 42. Esener N, Maciel-Guerra A, Giebel K, et al. Mass spectrometry and machine learning for the accurate diagnosis of benzylpenicillin and multidrug resistance of Staphylococcus aureus in bovine mastitis. PLOS Comput Biol. 2021;17(6):e1009108. CrossRef PubMed
- 43. Chen Y, Hong J, Chen Y, Wang H, Yu Y, Qu T. Characterization of a community-acquired methicillin-resistant sequence type 338 Staphylococcus aureus strain containing a staphylococcal cassette chromosome mec type VT. Int J Infect Dis. 2020;90:181-187. CrossRef PubMed
- 44. Maiden MCJ, Bygraves JA, Feil E, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95(6):3140-3145. CrossRef PubMed
- 45. Park SG, Lee HS, Park JY, Lee H. Molecular epidemiology of Staphylococcus aureus in skin and soft tissue infections and bone and joint infections in Korean children. J Korean Med Sci. 2019;34(49):e315. CrossRef PubMed
- 46. Votintseva AA, Fung R, Miller RR, et al. Prevalence of Staphylococcus aureus protein A (spa) mutants in the community and hospitals in Oxfordshire. BMC Microbiol. 2014;14(1):63. CrossRef PubMed
- 47. Keener AB, Thurlow LT, Kang S, et al. Staphylococcus aureus protein A disrupts immunity mediated by long-lived plasma cells. J Immunol. 2017;198(3):1263-1273. CrossRef PubMed
- 48. Goudarzi M, Fazeli M, Goudarzi H, Azad M, Seyedjavadi SS. Spa typing of Staphylococcus aureus strains isolated from clinical specimens of patients with nosocomial infections in Tehran, Iran. Jundishapur J Microbiol. 2016;9(7):e35685. CrossRef PubMed
- 49. Strommenger B, Braulke C, Heuck D, et al. Spa typing of Staphylococcus aureus as a frontline tool in epidemiological typing. J Clin Microbiol. 2008;46(2):574-581. CrossRef PubMed
- 50. Koreen L, Ramaswamy SV, Graviss EA, Naidich S, Musser JM, Kreiswirth BN. Spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J Clin Microbiol. 2004;42(2):792-799. CrossRef PubMed
- 51. Toleman MS, Reuter S, Jamrozy D, et al. Prospective genomic surveillance of methicillin-resistant Staphylococcus aureus (MRSA) associated with bloodstream infection, England, 1 October 2012 to 30 September 2013. Euro Surveill. 2019;24(4):1800215. CrossRef PubMed
- 52. Tani H, Miyata R, Ichikawa K, et al. Universal quenching probe system: flexible, specific, and cost-effective real-time polymerase chain reaction method. Anal Chem. 2009;81(14):5678-5685. CrossRef PubMed
- 53. Asadollahi P, Farahani NN, Mirzaii M, et al. Distribution of the most prevalent spa types among clinical isolates of methicillin-resistant and-susceptible Staphylococcus aureus around the world: a review. Front Microbiol. 2018;9:163. CrossRef PubMed
- 54. International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC). Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother. 2009;53:4961-4967. CrossRef PubMed
- 55. Ito T, Hiramatsu K, Tomasz A, et al; International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC). Guidelines for reporting novel mecA gene homologues. Antimicrob Agents Chemother. 2012;56(10):4997-4999. CrossRef PubMed
- 56. Chongtrakool P, Ito T, Ma XX, et al. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob Agents Chemother. 2006;50(3):1001-1012. CrossRef PubMed
- 57. Kaya H, Hasman H, Larsen J, et al. SCC mec Finder, a web-based tool for typing of staphylococcal cassette chromosome mec in Staphylococcus aureus using whole-genome sequence data. MSphere. 2018;3(1):e00612-e00617. CrossRef PubMed
- 58. Rajput A, Poudel S, Tsunemoto H, et al. Identifying the effect of vancomycin on health care-associated methicillin-resistant Staphylococcus aureus strains using bacteriological and physiological media. Gigascience. 2021;10(1):giaa156. CrossRef PubMed
- 59. Panwala T, Gandhi P, Jethwa D. Inducible Clindamycin resistance and MRSA amongst Staphylococcus aureus isolates: a phenotypic detection. IP Int J Med Microbiol Trop Dis 2020;6(4):222–226. CrossRef
- 60. Liu F, Rajabi S, Shi C, et al. Antibacterial activity of recently approved antibiotics against methicillin-resistant Staphylococcus aureus (MRSA) strains: a systematic review and meta-analysis. Ann Clin Microbiol Antimicrob. 2022;21(1):37. CrossRef PubMed
- 61. Kistler JM, Vroome CM, Ramsey FV, Ilyas AM. Increasing multidrug antibiotic resistance in MRSA infections of the hand: a 10-year analysis of risk factors. Hand (N Y). 2020;15(6):877-881. CrossRef PubMed
- 62. Kot B, Piechota M, Jakubczak A, et al. The prevalence of virulence determinants in methicillin-resistant Staphylococcus aureus isolated from different infections in hospitalized patients in Poland. Sci Rep. 2022;12(1):5477. CrossRef PubMed
- 63. Turner NA, Sharma-Kuinkel BK, Maskarinec SA, et al. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol. 2019;17(4):203-218. CrossRef PubMed
- 64. Otto M. MRSA virulence and spread. Cell Microbiol. 2012;14(10):1513-1521. CrossRef PubMed
- 65. Moormeier DE, Bayles KW. Staphylococcus aureus biofilm: a complex developmental organism. Mol Microbiol. 2017;104(3):365-376. CrossRef PubMed
- 66. Liu Y, Zhang J, Ji Y. Environmental factors modulate biofilm formation by Staphylococcus aureus. Sci Prog. 2020 Jan-Mar;103(1):36850419898659. CrossRef PubMed
- 67. Harraghy N, Hussain M, Haggar A, et al. The adhesive and immunomodulating properties of the multifunctional Staphylococcus aureus protein Eap. Microbiology (Reading). 2003;149(Pt 10):2701-2707. CrossRef PubMed
- 68. Siddiqui AH, Koirala J. Methicillin resistant Staphylococcus aureus. In: StatPearls. [internet] StatPearls Publishing 2021.
- 69. Tenover FC, Tickler IA, Le VM, Dewell S, Mendes RE, Goering RV. Updating molecular diagnostics for detecting methicillin-susceptible and methicillin-resistant Staphylococcus aureus isolates in blood culture bottles. J Clin Microbiol. 2019;57(11):e01195-e19. CrossRef PubMed
- 70. Martineau F, Picard FJ, Roy PH, Ouellette M, Bergeron MG. Species-specific and ubiquitous-DNA-based assays for rapid identification of Staphylococcus aureus. J Clin Microbiol. 1998;36(3):618-623. CrossRef PubMed
- 71. Tübbicke A, Hübner C, Hübner NO, Wegner C, Kramer A, Fleßa S. Cost comparison of MRSA screening and management – a decision tree analysis. BMC Health Serv Res. 2012;12(1):438. CrossRef PubMed
- 72. Brakstad OG, Aasbakk K, Maeland JA. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J Clin Microbiol. 1992;30(7):1654-1660. CrossRef PubMed
- 73. Chen C, Zhao Q, Guo J, Li Y, Chen Q. Identification of methicillin-resistant Staphylococcus aureus (MRSA) using simultaneous detection of mecA, nuc, and femB by Loop-Mediated Isothermal Amplification (LAMP). Curr Microbiol. 2017;74(8):965-971. CrossRef PubMed
- 74. Zhao L, Huang X, Zhang T, et al. A point-of-care test device for MRSA rapid detection. J Pharm Biomed Anal. 2022;209:114464. CrossRef PubMed
- 75. Uehara Y. Current status of Staphylococcal Cassette Chromosome mec (SCCmec). Antibiotics (Basel). 2022;11(1):86. CrossRef PubMed
- 76. Singh-Moodley A, Strasheim W, Mogokotleng R, Ismail H, Perovic O. Unconventional SCCmec types and low prevalence of the Panton-Valentine Leukocidin exotoxin in South African blood culture Staphylococcus aureus surveillance isolates, 2013-2016. PLoS One. 2019;14(11):e0225726. CrossRef PubMed
- 77. Burgold-Voigt S, Monecke S, Simbeck A, et al. Characterisation and molecular analysis of an unusual chimeric methicillin resistant Staphylococcus aureus strain and its bacteriophages. Front Genet. 2021;12:723958-723958. CrossRef PubMed
- 78. Kaku N, Sasaki D, Ota K, Miyazaki T, Yanagihara K. Changing molecular epidemiology and characteristics of MRSA isolated from bloodstream infections: nationwide surveillance in Japan in 2019. J Antimicrob Chemother. 2022;77(8):2130-2141. CrossRef PubMed
- 79. Lukassen MB, Saunders AM, Sindilariu P-D, Nielsen JL. Quantification of novel geosmin-producing bacteria in aquaculture systems. Aquaculture. 2017;479:304-310. CrossRef
- 80. Szabó J. Molecular methods in epidemiology of methicillin resistant Staphylococcus aureus (MRSA): advantages, disadvantages of different techniques. J Med Microbiol Diagn. 2014;3(3):1. CrossRef
- 81. Jaksik R, Iwanaszko M, Rzeszowska-Wolny J, Kimmel M. Microarray experiments and factors which affect their reliability. Biol Direct. 2015;10(1):46. CrossRef PubMed
