 |
Drug Target Insights 2020; 14: 16-25 DOI: 10.33393/dti.2020.2170 REVIEW |
 |
Prevalence of multidrug-resistant and extended-spectrum beta-lactamase (ESBL)-producing gram-negative bacilli: A meta-analysis report in Ethiopia
ABSTRACT
Multidrug-resistant (MDR) extended-spectrum beta-lactamase (ESBL)-producing bacterial isolates have emerged as a global threat to human health. Little is known about the overall prevalence of multidrug resistance profile and ESBL-producing gram-negative bacilli (GNB) in Ethiopia. Therefore, this meta-analysis was performed to produce proportional estimates of multidrug resistance and ESBL-producing GNB in Ethiopia.
A web-based search was conducted in PubMed, Google Scholar, Research Gate, Scopus and other databases. Articles published till 2019 on the prevalence and antimicrobial resistance profiles of ESBL-producing GNB in Ethiopia were included in the study. Relevant data were extracted and statistical analysis was performed using comprehensive meta-analysis version 3.3.0 software. Publication bias was analyzed and presented with funnel plots.
In this meta-analysis, the overall proportional estimate of ESBL-producing GNB was 48.9% (95% confidence interval [CI]: 0.402, 0.577). The pooled proportional estimates of ESBL-producing Klebsiella pneumoniae, Escherichia coli and other GNB were 61.8%, 41.2% and 42.9%, respectively. Regarding antimicrobial resistance profiles against selected drugs, the pooled proportional estimates of resistance against amoxicillin-clavulanic acid, trimethoprim-sulfamethoxazole, cefotaxime, ceftazidime, tetracycline, gentamicin and ciprofloxacin was 79.0%, 78.4%, 78.0%, 72.4%, 72.7%, 58.9% and 43.8%, respectively. The pooled proportional estimates of MDR isolates were found to be 82.7% (95% CI: 0.726, 0.896), which are relatively high as compared to other countries. This highlights a need for active surveillance systems which can help understand the actual epidemiology of ESBL, aid in formulating national guidelines for proper screening of ESBL and support developing standardized approaches for managing patients colonized with ESBL.
Keywords: Ethiopia, Extended-spectrum beta-lactamase, Gram negative, Multidrug resistance
Received: June 29, 2020
Accepted: September 14, 2020
Published online: October 5, 2020
© 2020 The Authors. This article is published by AboutScience and licensed under Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0). Any commercial use is not permitted and is subject to Publisher’s permissions. Full information is available at www.aboutscience.eu
Introduction
The production of extended-spectrum beta-lactamase (ESBL) enzymes is the main bacterial mechanism to acquire resistance to currently used broad-spectrum beta-lactam antibiotics. The infections caused by such enzyme-producing bacteria have significant potential impacts on antibiotic use and patient outcomes. Especially in resource-limited countries, limited availability of drugs coupled with inappropriate use could worsen the condition, increasing infections caused by such resistant strains, its emergency and transmission (1,2). For instance, in Ethiopia studies show that about 36.8% of the population got antibiotics from community drug retail outlets without a prescription and 67.9% of people had discontinued the use of antibiotics once their symptoms subside (3). Low educational status, dissatisfaction with healthcare services provided, pharmacy owners’ influence to maximize revenue, customer’s pressure, weak regulatory mechanism and professional conflicts of interest were found to be strong predictors of inappropriate use of antibiotic use among the community (3,4).
Enterobacteriaceae that produce ESBL carry plasmid-encoded enzymes that can efficiently hydrolyze and confer resistance to a variety of beta-lactam antibiotics, but not to carbapenems or cephamycins. Besides beta-lactam class of antibiotic, ESBL producers are commonly resistant to different families of antibiotics including fluoroquinolones, aminoglycosides and trimethoprim-sulfamethoxazole (SXT), which contribute to the selection and persistence of multidrug-resistant (MDR) ESBL strains and plasmids in both clinical and community settings. These enzymes are predominantly found in Escherichia coli and Klebsiella pneumoniae, although they are also present in other members of the gram-negative bacilli (GNB) (5,6,7).
ESBL-producing organisms play an important role in healthcare infections, increasing hospitalization time and morbidity and mortality rates (8). The presence of ESBL complicates antibiotic selection, especially in patients with serious infections, such as bacteremia. The reason for this is that ESBL-producing bacteria are often multiresistant to various antibiotics, an interesting feature of CTX-M (CTX stands for cefotaxime and M for Munich)-producing isolates is the coresistance to various classes of antibiotics such as fluoroquinolones, aminoglycosides and co-trimoxazole due to associated resistance mechanisms, which may be either chromosomally or plasmid-encoded (8,9,10,11).
The spread and the burden of ESBL-producing bacteria are greater in developing countries. For instance, the major epicenters of ESBL-expressing bacteria are located in Asia, Africa and the Middle East (12,13,14). Findings of a recent review showed that pooled prevalence of healthcare-associated infections in resource-limited settings (15.5%) was twice the average prevalence in Europe (7.1%) (15). Some plausible reasons for this difference include the following conditions that are prevalent in low-income countries: crowded hospitals, more extensive self-treatment and use of nonprescription antimicrobials, poorer hygiene in general and particularly in hospitals as well as less effective infection control.
Comprehensive data regarding ESBL-producing bacteria are generally lacking in African countries, compared with the developed world. In Ethiopia also, it is difficult to evaluate the spread and the burden of ESBL-producing organisms, because of the limited scope of studies and lack of coordinated epidemiological surveillance systems. By combining information from all relevant studies, meta-analyses can provide more precise estimates of the effects of healthcare than those derived from the individual studies included within a review. Therefore, to gain a better insight into the proportional estimates of MDR profiles and ESBL-producing GNB in Ethiopia, we retrieved available articles and collated the information in this study article.
Methods
Study design
A descriptive meta-analysis study comprising different studies on the prevalence and antimicrobial resistance profile of ESBL-producing GNB in Ethiopia was conducted.
Data sources and search strategies
A systematic and comprehensive search on available records up to December 2019 was carried out in PubMed, Google Scholar, Research Gate, Scopus and other databases. The following medical subject headings (MESH) in the title or abstract, such as “clinical specimen,” “ESBL,” OR “extended spectrum β-lactamase” “Enterobacteriaceae” OR “extended-spectrum-beta-lactamase,” “gram negatives” OR “antibacterial resistance,” “gram negatives” OR “antimicrobial susceptibility” AND “gram negatives” and “Ethiopia,” were searched. In addition, bibliographies of eligible studies and other meta-analyses were manually searched carefully to identify additional relevant articles.
Study selection and eligibility criteria
All articles related to the detection of ESBL-producing GNB from clinical specimens and hospital environment samples, written in English language, possessing approved microbiological methods for pathogen detection and containing sufficient and extractable data were included in the meta-analysis. Having assessed all the information from the recovered publications, online records that were available up to 2019 were considered as appropriate for eligibility assessment. All review articles and original articles conducted outside Ethiopia, articles with irretrievable full texts and records with unrelated outcomes of interest were excluded during screening and eligibility assessment.
Screening and eligibility criteria
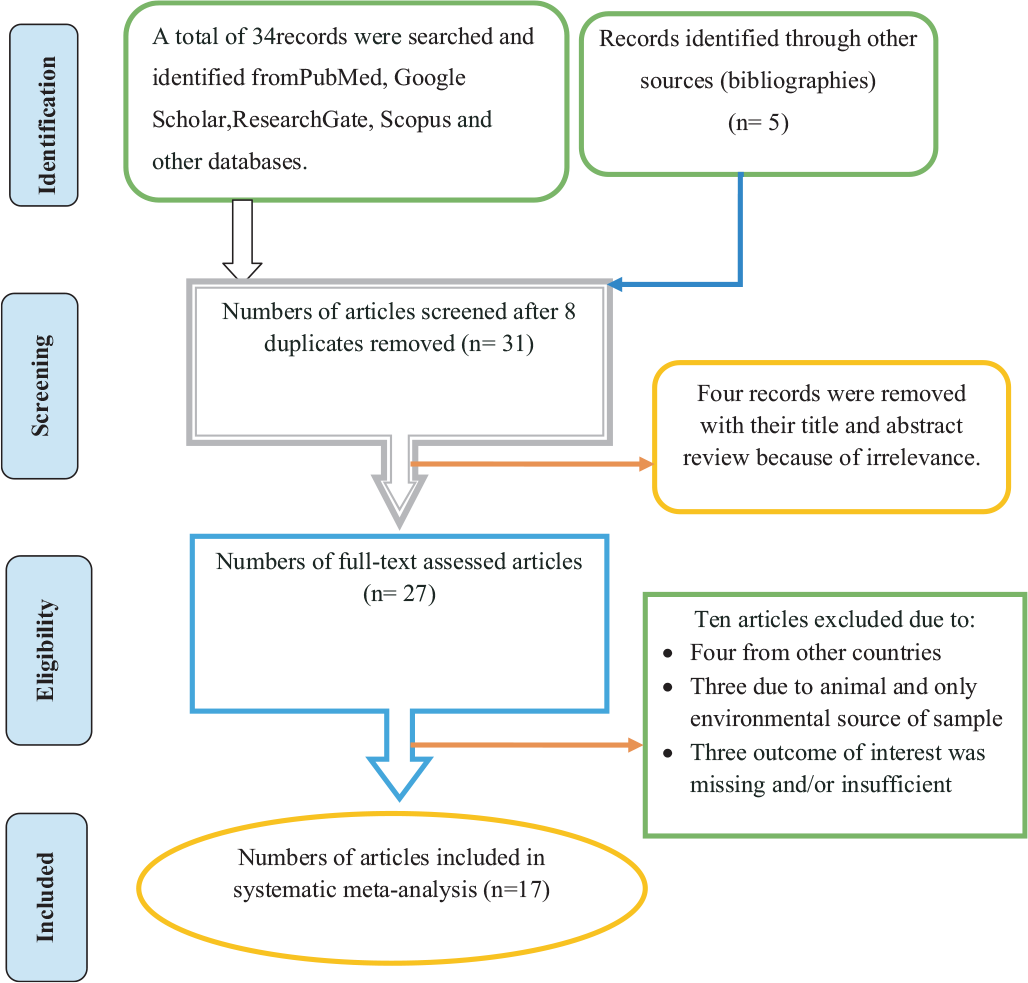
There were no limits with regard to study type except that the study had to be a primary study. However, the analysis was made to those studies which reported sufficient information to meet outcomes of interests. Some duplicates records were addressed manually due to variation in reference styles across sources. Thereafter, the authors (MA and TW) independently inspected all the titles and abstracts of articles related to the study question and these were included in a group of eligible articles with their own code and with irrelevant articles being excluded. All articles in the initially selected group were further screened in a second step by reviewing the full texts and evaluated for eligibility for final inclusion (Fig. 1).
Data extraction
Studies were identified by the main study name/identifier, followed by the year of publication. Using a predetermined, standardized and piloted data extraction form, the authors (MA and TW) independently extracted important data related to study characteristics from each article. Data extraction sheets were individually designed and pilot-tested using Microsoft Excel 2007. For each study meeting the review inclusion criteria, the following data, such as first author, study area, year of publication, study design, sample type, sample size, target population, isolate sources and outcomes of interests, number and common species isolated, proportion of ESBL-positive strains and proportion and/or number of drug and MDR isolate and the methods used to test for ESBL producers, were recorded. Data were extracted and analyzed at least twice to remove any discordance. Whenever there was discordance in the data extracted, a third person, a trained MSc laboratory professional, played a role in checking the data and a consensus was reached by double-checking of the articles with the two authors.
Quality assessment of included studies
The qualities of eligible studies were checked against items included in strengthening the reporting of observational studies in epidemiology (STROBE) Statement checklist (16). The quality scores (proportion) for each study were calculated against items of the STROBE checklist adequately. There were no limits with regard to study type except that the study had to be a primary study and we did not exclude any studies based on quality.
Outcome measurements
The first outcome measure is the proportion of ESBL-producing GNB from clinical specimens in Ethiopia. The pooled proportion of ESBL-producing GNB was calculated per bacterium isolate. The second outcome measure is the antimicrobial and MDR profiles of ESBL-producing and non-ESBL-producing GNB against selected antimicrobials of different categories.
Data processing and analysis
The relevant data were analyzed using comprehensive meta-analysis version 3.3.0 software (www.Meta-analysis.com). Both random and fixed effects models were used to calculate the pooled proportional estimates of ESBL-positive and MDR isolates. The I2 statistics was used to expresses the percentage of total variation across studies and significant heterogeneity was considered at p < 0.05 and I2 > 50%. Publication bias was evaluated by using Begg’s and Egger’s tests and presented with funnel plots of standard error of Logit event rate, and a statistical significant publication bias was considered at p < 0.05.
Results
Search and screening results and distribution of included articles
A total of 39 studies were identified from several sources including PubMed, Google Scholar, Research Gate, Scopus and other databases. After removing eight duplicated articles, 31 were screened and four records were removed with their title and abstract review because of irrelevance. After full-text assessment, ten articles excluded: four from other countries, three due to animal and only nonhospital environment source of sample and three outcome of interest was missing and/or insufficient. Seventeen articles fulfilling the eligibility criteria were included for systematic meta-analysis (Fig. 1). The eligible studies were published in the year up to 2019. The study design of all included articles was cross-sectional studies. Most of the studies indicated that various specimens had been utilized for screening of GNB; particularly biological fluids like blood, urine, pus, stool and cerebrospinal fluid (CSF) were taken for test. Hospital environment samples such as wastewater and different swab samples from hospital contact surfaces were also taken for test. With regard to sources of biological samples, most of the studies included inpatients and outpatients as their sources of samples. With regard to study areas, six (35.3%) were conducted in the southwestern parts of Ethiopia, six (35.3%) in central Ethiopia, four (23.5%) in northern parts of Ethiopia and one (5.9%) in eastern parts of Ethiopia. Based on the available evidence, the maximum number of articles on this subject was published in the year 2014, which means only three articles were published in 2005, 2011 and 2012 (Tab. I). The median score of published studies against STROBE items was 72.2% (with a range of [50.7-79.7]).
Proportional estimates of ESBL-positive GNB
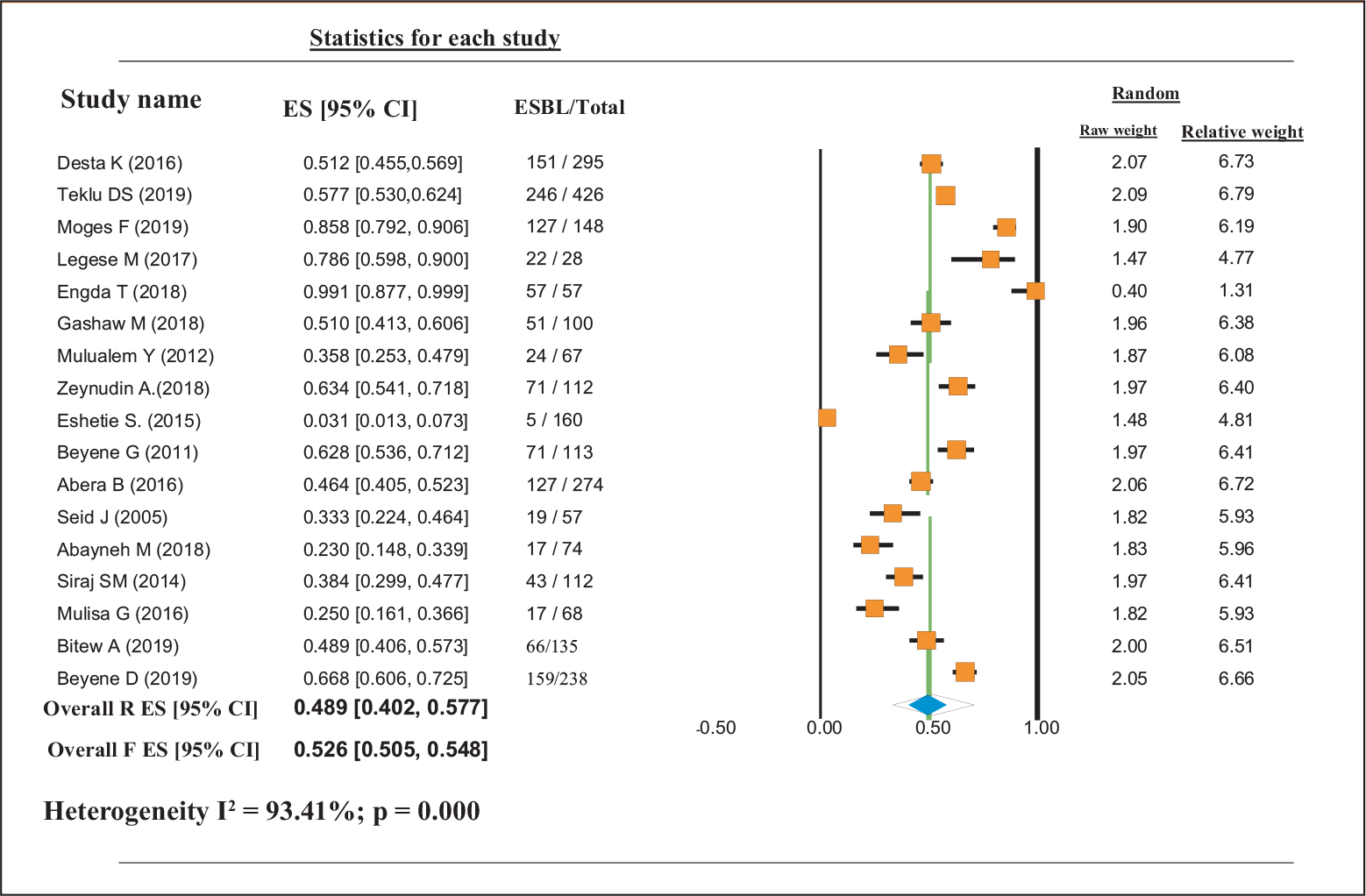
This study indicates that although different bacterial pathogens have the probability of producing ESBL as their resistance mechanisms, studies on the proportion of ESBL-producing bacteria have not been carried out properly in all parts of Ethiopia. Based on the available data, the pooled proportional estimate of ESBL-producing GNB in Ethiopia was 0.489 (95% confidence interval [CI]: 0.402, 0.577). Proportional estimates of ESBL-producing K. pneumoniae were 0.618 (95% CI: 0.487, 0.734), whereas proportional estimates of ESBL-producing E. coli were 0.412 (95% CI: 0.326, 0.504). The proportional estimate of other ESBL-producing GNB was 0.429 (95% CI: 0.352, 0.509). Overall heterogeneity was significant [I2 = 93.41%; p = 0.000] (Tab. II and Fig. 2).
Proportional estimates of MDR profiles of bacterial isolates
The bacterial isolates that showed different antimicrobial resistance profile against selected agents were extracted. Accordingly, the pooled estimate indicated that 0.720 (95% CI: 0.586, 0.820) and 0.780 (95% CI: 0.615, 0.891) of the ESBL-producing and non-ESBL-producing GNB were found resistant to third-generation cephalosporin (ceftazidime and cefotaxime), respectively. The pooled estimate indicated that 0.589 (95% CI: 0.482, 0.688), 0.438 (95% CI: 0.347, 0.534) and 0.790 (95% CI: 0.713, 0.850) of ESBL-producing and non-ESBL-producing GNB isolates were resistant to gentamicin, ciprofloxacin and amoxicillin-clavulanic acid, respectively. Besides, 0.784 (95% CI: 0.726, 0.832) and 0.727 (95% CI: 0.605, 0.823) of ESBL-producing and non-ESBL-producing GNB isolates were resistant to SXT and tetracycline (TET), respectively. The pooled proportional estimates of MDR isolates were found to be 0.827 (95% CI: 0.726, 0.896) (Tab. III).
| First author, publication year | Study design | Study area | Sample types | Sample sources | Sample size | Total isolates | Total ESBL positives | Common ESBL producers | ESBL confirmationmethod | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| E. coli | K. pneumoniae | Other gram-negatives | |||||||||
| Desta et al, 2016 (17) | CS | AA | Fecal sample/swab | IP | 267 | 295 | 151/295 | 106/235 | 44/58 | 1/2 | CDT + Vitek2 |
| Teklu et al, 2019 (18) | CS | AA | Urine, blood, sputum, CSF, body fluid, pus and discharge | IP and OP | 426 | 426 | 246/426 | 119/228 | 81/103 | 46/95 | CDT |
| Moges et al, 2019 (19) | CS | Bahir Dar | Urine, blood, stool, pus, sputum, CSF, body fluid, ear, nasal, cervical discharge | IP and OP | 532 | 263 | 127/148 | 14/23 | 79/97 | 34/76 | CDT |
| Legese et al, 2017 (20) | CS | AA | Blood and urine | IP and OP | 322 | 33 | 22/28 | 5/6 | 16/19 | 1/8 | CDT |
| Engda et al, 2018 (21) | CS | Gondar | Swabs of sinks, bed, door handles, wastewater | Hospital env’t | 384 | 57 | 57/57 | 20/57 | 24/57 | 13/57 | Hicrome ESBL agar |
| Gashaw et al, 2018 (22) | CS | Jimma | Urine, blood, sputum, wound/pus swab | IP | 118 | 126 | 51/100 | 19/31 | 16/30 | 18/39 | DDST |
| Mulualem et al, 2012 (23) | CS | Jimma | Urine, stool, sputum, wound/pus swab | IP and OP | 359 | 67 | 24/67 | 24/67 | NA | NA | CDT |
| Zeynudin et al, 2018 (24) | CS | Jimma | Wound swabs, urine, biopsies, sputum | IP and OP | 224 | 224 | 71/112 | 13/13 | 30/31 | 28/68 | Check-MDR Microarray Kits |
| Eshetie et al, 2015 (25) | CS | Gondar | Urine | IP and OP | 442 | 183 | 5/160 | 2/104 | 3/28 | 42 (NA) | CHROMagar |
| Beyene et al, 2011 (26) | CS | Jimma | Stool, blood | IP and OP | 1,225 | 113 | 71/113 | NA | NA | 71/113 | E-test |
| Abera et al, 2016 (27) | CS | Bahir Dar | Blood, urine, pus, CSF, ear discharges, wound swab, water | IP and OP | 757 | 274 | 127/274 | 73/170 | 36/55 | 17/49 | DDST |
| Seid et al, 2005 (28) | CS | Harrar | Sputum, urine and pus | IP and OP | 384 | 57 | 19/57 | NA | 19/57 | NA | CDT |
| Abayneh et al., 2018 (29) | CS | Jimma | Urine | OP | 342 | 74 | 17/74 | 13/63 | 4/11 | NA | DDST |
| Siraj et al, 2014 (30) | CS | Jimma | Urine, sputum, blood, vaginal swabs, wound/pus swab, eye discharge | IP and OP | 471 | 112 | 43/112 | 24/85 | 19/27 | NA | DDST |
| Mulisa et al, 2016 (31) | CS | Adama | Urine, wound, nasal, stool, pleural fluids, ear discharge | IP and OP | 384 | 133 | 17/68 | 10/35 | 2/8 | 5/25 | DDST |
| Bitew (2019) (32) | CS | AA | Urine, wound, blood, CSF, ear, nasal | IP and OP | 996 | 153 | 66/135 | NA | NA | 66/135 | CDT |
| Beyene (2019) (33) | CS | AA | Urine, sputum, blood, vaginal, wound/pus, eye discharge | IP | 947 | 238 | 159/238 | 91/144 | 55/72 | 13/21 | CDT |
AA = Addis Ababa; CDT = Combination Disc Test; CS = cross-sectional; CSF = cerebrospinal fluid; DDST = Double Disc Synergy Test; ESBL = extended-spectrum beta-lactamase; E-Test = Epsilometric Test; IP = inpatient; MDR = multidrug resistance; NA = not analyzed; OP = outpatient.
| Studies | Total proportions of ESBL | Proportions of ESBL-producing E. coli | Proportions of ESBL-producing K. pneumoniae | Proportions of ESBL-producing other GNs | ||||
|---|---|---|---|---|---|---|---|---|
| ES [95% CI] | Weight | ES [95% CI] | Weight | ES [95% CI] | Weight | ES [95% CI] | Weight | |
| Desta et al (2016) | 0.51 [0.455, 0.569] | 1.80 | 0.451 [0.389, 0.515] | 2.91 | 0.759 [0.633, 0.852] | 1.00 | 0.500 [0.059, 0.941] | 0.45 |
| Teklu et al (2019) | 0.58 [0.530, 0.624] | 1.81 | 0.522 [0.457, 0.586] | 2.91 | 0.786 [0.697, 0.855] | 1.04 | 0.484 [0.386, 0.584] | 3.53 |
| Moges et al (2019) | 0.858 [0.792, 0.906] | 1.67 | 0.609 [0.402, 0.782] | 1.96 | 0.814 [0.725, 0.880] | 1.03 | 0.447 [0.340, 0.560] | 3.40 |
| Legese et al (2017) | 0.786 [0.598, 0.900] | 1.32 | 0.833 [0.369, 0.977] | 0.66 | 0.842 [0.608, 0.948] | 0.77 | 0.125 [0.017, 0.537] | 0.72 |
| Engda et al (2018) | 0.991 [0.877, 0.999] | 0.39 | 0.351 [0.239, 0.482] | 2.48 | 0.421 [0.301, 0.552] | 1.02 | 0.228 [0.137, 0.354] | 2.93 |
| Gashaw et al (2018) | 0.510 [0.413, 0.606] | 1.71 | 0.613 [0.435, 0.765] | 2.16 | 0.533 [0.358, 0.701] | 0.96 | 0.462 [0.317, 0.617] | 2.90 |
| Mulualem et al (2012) | 0.358 [0.253, 0.479] | 1.64 | 0.358 [0.253, 0.479] | 2.55 | NA | NA | NA | |
| Zeynudin et al (2018) | 0.634 [0.541, 0.718] | 1.72 | 0.964 [0.616, 0.998] | 0.42 | 0.968 [0.804, 0.995] | 0.52 | 0.412 [0.302, 0.532] | 3.31 |
| Eshetie et al (2015) | 0.027 [0.011, 0.064] | 1.34 | 0.018 [0.004, 0.069] | 1.20 | 0.103 [0.034, 0.276] | 0.78 | 42 (NA) | NA |
| Beyene et al (2011) | 0.628 [0.536, 0.712] | 1.72 | NA | NA | NA | 0.628 [0.536, 0.712] | 3.58 | |
| Abera et al (2016) | 0.464 [0.405, 0.523] | 1.79 | 0.429 [0.357, 0.505] | 2.85 | 0.655 [0.521, 0.768] | 1.01 | 0.415 [0.276, 0.569] | 2.93 |
| Seid et al (2005) | 0.333 [0.224, 0.464] | 1.61 | NA | NA | 0.333 [0.224, 0.464] | 1.02 | NA | NA |
| Abayneh et al (2018) | 0.230 [0.148, 0.339] | 1.61 | 0.206 [0.124, 0.324] | 2.36 | 0.364 [0.143, 0.661] | 0.77 | NA | NA |
| Siraj et al (2014) | 0.384 [0.299, 0.477] | 1.72 | 0.282 [0.197, 0.387] | 2.60 | 0.704 [0.510, 0.844] | 0.92 | NA | NA |
| Mulisa et al (2016) | 0.250 [0.161, 0.366] | 1.61 | 0.286 [0.161, 0.454] | 2.14 | 0.250 [ 0.063, 0.623] | 0.64 | 0.200 [0.086, 0.400] | 2.04 |
| Bitew (2019) | 0.489 [0.406, 0.573] | 2.00 | NA | NA | NA | 0.489 [0.406, 0.573] | 4.42 | |
| Beyene (2019) | 0.668 [0.606, 0.725] | 20.5 | 0.632 [0.550, 0.707] | 2.57 | 0.764 [0.653, 0.848] | 1.11 | 0.619 [0.402, 0.797] | 2.51 |
| Overall R ES [95% CI] | 0.489 [0.402, 0.577] | 0.412 [0.326, 0.504] | 0.618 [0.487, 0.734] | 0.429 [0.352, 0.509] | ||||
| Overall F ES [95% CI] | 0.526 [0.505, 0.548] | 0.451 [0.422, 0.481] | 0.646 [0.604, 0.686] | 0.461 [0.424, 0.500] | ||||
CI = confidence interval; ESBL = extended-spectrum beta-lactamase; F = fixed; GN = gram negatives; NA = not analyzed; R = random; ES = event rate.
| Authors | Total isolates | Total ESBL-producer | Antimicrobial resistance profiles | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CAZ | CTX | GNT | CIP | SXT | TET | AMC | MDR | |||
| Desta et al. (2016) | 295 | 151/295 | 137/150 | 146/150 | 105/150 | 94/150 | 136/150 | NA | 134/150 | 150/150 |
| Teklu et al. (2019) | 426 | 246/426 | 257/426 | 265/426 | 185/426 | 240/426 | 324/426 | NA | 305/426 | 237/246 or 291/426* |
| Moges et al. (2019) | 185 | 127/148 | 143/148 | 129/148 | 116/148 | 52/148 | 138/148 | 127/148 | 117/148 | 127/148 or 148/185* |
| Legese et al. (2017) | 33 | 22/28 | NA | 29/33 | 26/33 | NA | 29/33 | 25/33 | 28/33 | NA |
| Engda et al. (2018) | 57 | 57/57 | 57/57 | 57/57 | 11/57 | 25/57 | 37/57 | NA | 57/57 | 32/57 |
| Gashaw et al. (2018) | 100 | 51/100 | 63/100 | 60/100 | 68/100 | 48/100 | 79/100 | 90/100 | 94/100 | 28/100* |
| Mulualem et al. (2012) | 67 | 24/67 | 4/67 | 6/67 | 2/67 | 14/67 | 38/67 | 49/67 | 47/67 | 67/67* |
| Zeynudin et al. (2018) | 224 | 71/112 | 63/68 | 66/68 | 60/68 | 41/68 | 62/68 | NA | NA | 67/68* |
| Eshetie et al. (2015) | 183 | 5/183 | 90/160 | 40/160 | 94/160 | 4/160 | 103/160 | 79/160 | 81/160 | 160/183* |
| Beyene et al. (2011) | 113 | 71/113 | NA | NA | 84 | 1 | 91 | 45 | NA | 78/113* |
| Abera et al. (2016) | 274 | 127/274 | NA | NA | NA | 106/274 | 174/274 | NA | NA | NA |
| Seid et al. (2005) | 57 | 19/57 | 23/57 | 22/57 | 35/57 | NA | 37/57 | NA | NA | 41/57* |
| Abayneh et al. (2018) | 74 | 17/74 | 12/17 | 17/17 | 11/17 | 13/17 | 14/17 | 14/17 | 14/17 | 14/17 |
| Siraj et al. (2014) | 112 | 43/112 | 42/43 | 43/43 | 36/43 | 33/43 | 41/43 | 39/43 | 38/43 | 38/43 |
| Mulisa et al. (2016) | 68 | 17/68 | NA | NA | 12/17 | 14/17 | 14/17 | 6/17 | NA | 17/17 |
| Bitew (2019) | 135 | 66/135 | 45/135 | NA | 32/135 | 47/135 | 86/135 | 92/135 | 100/135 | 110/135* |
| Beyene (2019) | 238 | 159/238 | 176/238 | NA | 117/238 | 137/238 | 195/238 | 191/238 | 148/238 | 225/238* |
| Overall R, ES [95% CI] | 1273/2487 | 0.72 [0.586, 0.820] | 0.78 [0.615, 0.891] | 0.589 [0.482, 0.688] | 0.438 [0.347, 0.534] | 0.784 [0.726, 0.832] | 0.727 [0.605, 0.823] | 0.790 [0.713, 0.850] | 0.827 [0.726, 0.896] | |
AMC = ampicillin-clavulanic acid; CAZ = ceftazidime; CI = confidence interval; CIP = ciprofloxacin; CTX = cefotaxime; GNT = gentamicin; ESBL = extended-spectrum beta-lactamase; MDR = multi-drug resistance; NA = not analyzed; R = resistance; SXT = trimethoprim-sulfamethoxazole; TET = tetracycline; ES = event rate.
*From overall isolates.
Laboratory methods used to estimate the proportion of ESBL-producing strains
According to this review, 8/17 (47.1%) of the articles reviewed used combination disk test (CDT) and 5/17 (29.4%) articles used double disk synergy test (DDST) methods alone. Two (11.8%) articles used CHROMagar and one article used E-test to estimate ESBL proportions. Only one article used Check-MDR CT103 Microarray Kits for detection and molecular characterization of the ESBL genes (Tab. I).
Publication bias
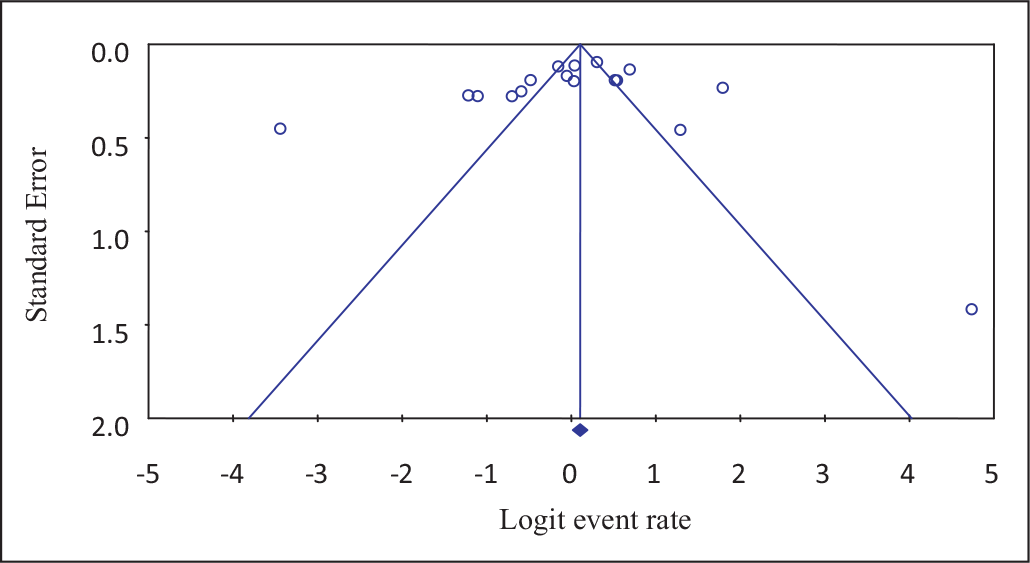
Funnel plots of standard error with Logit event rate confirmed that there is no statistically significant evidence of publication bias on studies reporting the prevalence of ESBL-producing bacterial isolates from different clinical samples in Ethiopia (Begg’s test, p = 0.217; Egger’s test, p = 0.231) (Fig. 3). However, Begg suggested that a nonsignificant correlation may be due to low statistical power and cannot be taken as evidence that bias is absent.
Discussion
This is the first meta-analysis study relating the extent of the ESBL-producing GNB in Ethiopia. Accordingly, the pooled proportional estimates of ESBL-producing GNB were 48.9%, which is higher than previous estimates of meta-analysis in East Africa (34) and Pakistan (35), in which overall pooled proportion of ESBL-producers was 42% and 40%, respectively. As compared to each country in East Africa, this finding is higher than the pooled proportion of ESBL-producing Enterobacteriaceae in Tanzania (39%) and Kenya (47%) (34). The result is also higher than estimates of meta-analysis in African countries, in which the total proportion of ESBL-producing isolates was 15% in 16 out of 26 studies (36).
As compared to resource-rich countries, the pooled proportion of ESBL-producing isolates was thus considerably higher in resource-limited countries, including our setting. For instance, the pooled global prevalence of ESBL‐producing Enterobacteriaceae among pregnant women diagnosed with urinary tract infections (UTIs) is 25%, with the highest rates in Africa (45%) and India (33%), followed by 15% in other Asian countries, 5% (2.8%) in Europe and the lowest one of 4% in South America and 3% in North America (37). This study finding is also close to data reported for China, where a nationwide survey that included 30 hospitals reported over 46% resistance due to ESBL (38). However, our estimate is slightly lower than for Uganda (62%) (34), Ghana (49%) (39), Cameroon (54%) (40) and Morocco (43%) (41).
With regard to the frequency of isolates, in this study, the pooled proportional estimates of ESBL-producing K. pneumoniae, E. coli and other GNB were 60.3%, 39.0% and 40.9%, respectively. This is much higher than a SMART study between 2009 and 2010, in which ESBL prevalence among K. pneumoniae and E. coli was 38.9% and 17.6%, respectively, in Europe, and 8.8% and 8.5, respectively, in North America (42). Moreover, our meta-analysis result is also higher than a report of in vitro activity of tigecycline and comparators against gram-negative and gram-positive organisms collected from Asia-Pacific during 2004-2010 and 2015, in which 24.6% and 15.8% of E. coli and K. pneumoniae were ESBL producers in 2015, respectively[A: Please check the clarity of this sentence.] (43). In addition, among GNB collected from intra-abdominal infections in the Asia-Pacific region during 2007, 42.2% and 35.8% of E. coli and Klebsiella spp., respectively, were ESBL positive (44). However, higher proportions of ESBL-producing E. coli and K. pneumoniae were reported from India, Vietnam and China (44,45).
In this study, inter-study results showed a wide and statistically significant degree of variation in proportion estimates of ESBL proportions (p < 0.05). There are several possible factors that may account for the variations seen in this review. The first factor is the difference in sensitivity and specificity between methods used in estimating ESBL proportions. Some studies reviewed estimated ESBL proportions using purely phenotypic methods, while others used both phenotypic and molecular-based methods. For instance, in this study, most of the reviewed articles used CDT (17,18,19,20,23,28,32,33) and DDST (22, 27,29,30,31) methods alone for the detection of ESBL-producing isolates, and only one study used molecular technique for investigations and characterizations of the gene encoding ESBL (24).
The other factor contributing to the variation in proportion estimates of ESBL proportions is type of wards or units, type of specimen collected and whether patients were attending outpatient or inpatient departments. In this meta-analysis, many reviewed studies showed that ESBL-producing isolates were more among inpatients than outpatients (20,24,30). In contrast, two reviewed studies showed the reverse finding (23,27). A meta-analysis study in areas of sub-Saharan Africa showed that pooled ESBL-producing Enterobacteriaceae colonization was 18%, 32% and 55% in community studies, at hospital admission and for inpatient studies, respectively (46). Many reports have documented the difference in ESBL proportion estimates between hospitals versus community-based surveys (23,25,27,30). However, the lack of any estimates for community-based ESBL carriage in Ethiopia underscores an urgent need for surveillance in the region. Infection control in hospitals including hand hygiene and rational antibiotic use can be effective measures to stop further spread of the ESBL-producing and MDR strains in both hospitals and communities.
MDR GNB are now globally widespread and present a major challenge to modern medical practice. To date, the overall epidemiology and burden of MDR bacteria and their mechanisms of resistance have not been fully understood, especially in resource-limited countries, including Ethiopia. This is the first meta-analysis study conducted to determine the pooled proportional estimates of ESBL-producing GNB and MDR isolates in Ethiopia. Accordingly, the pooled proportional estimates of MDR isolates were found to be 82.7% (Tab. III). This finding is higher than a finding of previous meta-analysis in Ethiopia, in which the pooled proportional estimates of MDR isolates were 59.7% (47); however, it was lower than a single study finding in Ethiopia (33) and Chile (48), in which 94.5% and 100% isolates exhibited MDR profiles. Lower study finding was also reported in Ethiopia, in which the overall prevalence of MDR was 69.9%, of which 81.5% was in the hospital environment, while 54.2% was in non-hospital environment samples (49). The occurrence of MDR may be linked with indiscriminate utilization of antimicrobial agents such as wrong indication, wrong duration, improper route of administration, use of leftover antibiotics from a family member and improper discontinuation of antibiotics or genetic mutation (47,50,51). A meta-analysis finding in Ethiopia indicated that the pooled estimate of inappropriate antibiotic use was 49.2% and the pooled proportion of self-antibiotic prescription was 43.3% (47). Thus, the frequent and inappropriate use of antibiotics in humans and animals may contribute to the recent emergence of ESBL producers and MDR strains both in healthcare institutions and communities.
Moreover, medication required to treat ESBL-producing isolates is expensive and unaffordable for the majority of the population in these settings, making these bacteria difficult to treat. A study in Ethiopia concluded that mortality was significantly associated with antimicrobial resistance (52). For instance, all 11 patients with Enterobacteriaceae resistant to third-generation cephalosporins died. Another systematic review and meta-analysis estimated that the mortality in neonates with bloodstream infections (BSIs) due to ESBL-producing Enterobacteriaceae was 36%, as compared to 18% among all other neonates with BSI (53). It is therefore of the utmost importance to make reliable data available to guide strategies devoted to limiting the spread of particularly ESBL-producing pathogens and MDR strains in Ethiopia.
In this study, significant numbers of bacterial isolates showed different resistance profile against selected antimicrobial agents. The pooled proportional estimate of resistance against third-generation cephalosporins (cefotaxime and ceftazidime), amoxicillin-clavulanic acid, gentamicin, ciprofloxacin, SXT and TET ranges from 43.8% to 79.0%. Almost similar findings were reported in Ethiopia and Chile, in which 45.4% to >88% isolates exhibit resistance against beta-lactams and non-lactam drugs (33,48). This high resistance rate might be associated with an interesting feature of ESBL-producing isolates as resistance mechanisms, which may be either chromosomally or plasmid-encoded, especially CTX-M that often showed feature of coresistance to various classes of antibiotics such as fluoroquinolones, aminoglycosides, and co-trimoxazole. Therefore, interventions including infection control measures and restriction of low-quality and inappropriate use of antibiotics may aid in controlling the emergency and spread of ESBL-producing pathogens and may actually prove cost-beneficial. As a limitation, in this study, due to the lack of sufficient records on the prevalence of ESBL producers from different clinical samples in Ethiopia, all primary studies, including those with very small datasets (<10 ESBL-producing strains) were included.
Conclusion
The problems related to antibiotic resistance, including MDR due to ESBL, are significant in Ethiopia. The scarcity of data on predictors, clinical outcomes, magnitudes and gene variants encoding resistance due to ESBL-producing GNB calls for active surveillance systems, which can help understand the current epidemiology of ESBL within the country. Furthermore, this can aid in developing national guidelines for proper screening of ESBL as well as developing standardized approaches for managing patients colonized with ESBL-producing GNB.
Disclosures
Conflict of interest: The authors declare that they have no competing interest.
Financial support: No funding was allocated for this study.
Availability of data and materials: All the data supporting our findings were incorporated within the manuscript, but are available from the corresponding author on reasonable request.
Authors’ contributions: MA and TW participated in the study design, study searching and screening and data extractions. MA analyzed the data and drafted the manuscript. TW support data analysis, read, revised and approved the final version of the manuscript.
References
- 1. Dhillon RH, Clark J. ESBLs: a clear and present danger? Hindawi Publ Corp. 2012;11.
- 2. Byarugaba DK. Antimicrobial resistance in developing countries Springer, 2009;15-27.
- 3. Erku DA, Mekuria AB, Belachew SA. Inappropriate use of antibiotics among communities of Gondar town, Ethiopia: a threat to the development of antimicrobial resistance. Antimicrob Resist Infect Control. 2017;6:112. Crossref
- 4. Gebretekle GB, Serbessa MK. Exploration of over the counter sales of antibiotics in community pharmacies of Addis Ababa, Ethiopia: pharmacy professionals’ perspective. Antimicrob Resist Infect Control. 2016;5:2. Crossref
- 5. European Centre for Disease Prevention and Control. Systematic review of the effectiveness of infection control measures to prevent the transmission of extended-spectrum beta-lactamase-producing Enterobacteriaceae through cross-border transfer of patients. Stockholm: ECDC 2014. Crossref
- 6. Morosini MI, García-Castillo M, Coque TM, et al. Antibiotic co-resistance in ESBL-producing Enterobacteriaceae and in vitro activity of tigecycline. Antimicrob Agents Chemother. 2006;50(8):2695-2699.
- 7. Cantón R, Novais A, Valverde A, et al. Prevalence and spread of extended-spectrum beta-lactamase-producing Enterobacteriaceae in Europe. Clin Microbiol Infect. 2008;14 Suppl 1:144-153.
- 8. Shamsrizi P, Gladstone BP, Carrara E, et al. Variation of effect estimates in the analysis of mortality and length of hospital stay in patients with infections caused by bacteria-producing extended-spectrum beta-lactamases: a systematic review and meta-analysis. BMJ Open. 2020;10:e030266. Crossref
- 9. Paterson DL, Bonomo RA. Extended-spectrum β-lactamases: a clinical update. Clin Microbiol Rev. 2005;18(4):657-686.
- 10. Giske CG, Monnet DL, Cars O, Carmeli Y, . Clinical and economic impact of common multidrug-resistant gram-negative bacilli. Antimicrob Agents Chemother. 2008;52(3):813-821
- 11. Bonnet R. Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother. 2004;48(1):1-14.
- 12. Villegas M V, Kattan JN, Quinteros MG, Casellas JM. Prevalence of extended-spectrum b-lactamases in South America. Clin Microbiol Infect. European Society of Clinical Microbiology and Infectious Diseases; 2008;14:154-158.
- 13. Hawkey PM. Prevalence and clonality of extended-spectrum beta-lactamases in Asia. Clin Microbiol Infect. 2008;14:159-165.
- 14. Bradford PA. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Am Soc Microbiol. 2001;14(4):933-951.
- 15. Tham J, Melander E, Walder M, Edquist PJ, Odenholt I. Prevalence of faecal ESBL carriage in the community and in a hospital setting in a county of Southern Sweden. Eur J Clin Microbiol Infect Dis. 2011;10:1159-1162
- 16. Malta M, Cardoso LO, Bastos FI, Magnanini MM, Silva CM. STROBE initiative: guidelines on reporting observational studies. Saude Publica. 2010;44(3):559-565. Crossref
- 17. Desta K, Woldeamanuel Y, Azazh A, et al. High gastrointestinal colonization rate with extended-spectrum β-lactamase-producing Enterobacteriaceae in hospitalized patients: emergence of carbapenemase-producing K. pneumoniae in Ethiopia. PLoS One. 2016;11(8):e0161685. Crossref
- 18. Teklu DS, Negeri AA, Legese MH, Bedada TL, Woldemariam HK, Tullu KD. Extended-spectrum beta-lactamase-production and multi-drug resistance among Enterobacteriaceae isolated in Addis Ababa, Ethiopia. Antimicrob Resist Infect Control. 2019; 8:39. Crossref
- 19. Moges F, Eshetie S, Abebe W, et al. High prevalence of extended-spectrum beta-lactamase-producing gram-negative pathogens from patients attending Felege Hiwot Comprehensive Specialized Hospital, Bahir Dar, Amhara region. PLoS One. 2019;14(4):e0215177. Crossref
- 20. Legese MH, Weldearegay MG, Asrat D. Extended-spectrum beta-lactamase- and carbapenemase-producing Enterobacteriaceae among Ethiopian children. Infect Drug Resist. 2017;10:27-34. Crossref
- 21. Engda T, Moges F, Gelaw A, Eshete S, Mekonnen F. Prevalence and antimicrobial susceptibility patterns of extended spectrum beta-lactamase producing Entrobacteriaceae in the University of Gondar Referral Hospital environments, northwest Ethiopia. BMC Res Notes. 2018;11:335. Crossref
- 22. Gashaw M, Berhane M, Bekele S, et al. Emergence of high drug resistant bacterial isolates from patients with health care associated infections at Jimma University medical center: a cross sectional study. Antimicrob Resist Infect Control. 2018;7:138. Crossref
- 23. Mulualem Y, Kasa T, Mekonnen Z, Suleman S. Occurrence of extended spectrum beta (beta)-lactamases in multi-drug resistant Escherichia coli isolated from a clinical setting in Jimma University Specialized Hospital, Jimma, and Southwest Ethiopia. East Afr J Public Health. 2012;9(2):58-61. PMID: 23139958.
- 24. Zeynudin A, Pritsch M, Schubert S, et al. Prevalence and antibiotic susceptibility pattern of CTX-M type extended-spectrum β-lactamases among clinical isolates of gram-negative bacilli in Jimma, Ethiopia. BMC Infect Dis.2018;18:524. Crossref
- 25. Eshetie S, Unakal C, Gelaw A, Ayelign B, Endris M, Moges F. Multidrug resistant and carbapenemase producing Enterobacteriaceae among patients with urinary tract infection at referral Hospital, Northwest Ethiopia. Antimicrob Resist Infect Control. 2015;4:12. Crossref
- 26. Beyene G, Nair S, Asrat D, Mengistu Y, Engers H, Wain J. Multidrug resistant Salmonella Concord is a major cause of salmonellosis in children in Ethiopia. J Infect DevCtries. 2011;5:023-033. Crossref
- 27. Abera B, Kibret M, Mulu W. Extended-spectrum beta (β)-lactamases and antibiogram in Enterobacteriaceae from clinical and drinking water sources from Bahir Dar City, Ethiopia. PLoS One. 2016;11(11):e0166519. Crossref
- 28. Seid J, Asrat D. Occurrence of extended spectrum beta-lactamse enzymes in clinical isolates of Klebsiella species from Harar region, eastern Ethiopia. Acta Trop. 2005;95(2);143-148. Crossref
- 29. Abayneh M, Tesfaw G, Abdissa A. Isolation of extended-spectrum β-lactamase-(ESBL) - producing E. coli and K. pneumoniae from patients with community-onset urinary tract infections in Jimma University Specialized Hospital, Southwest Ethiopia. Can J Infect Dis Med Microbiol. 2018;2018:4846159. Crossref
- 30. Siraj SM, Ali S, Wondafrash B. Extended-spectrum-lactamase production and antimicrobial resistance in Klebsiellapneumoniae and Escherichia coli among inpatients and outpatients of Jimma University Specialized Hospital, South-west, Ethiopia. Afr J Microbiol Res. 2014;8(43):3687-3694.
- 31. Mulisa G, Selassie L, Jarso G, Shiferew T, Zewdu A. Prevalence of extended spectrum beta-lactamase producing Enterobacteriaceae: a cross sectional study at Adama Hospital, Adama, Ethiopia. J Emerg Infect DisPatho. 2016;1(1):102.
- 32. Bitew A. High prevalence of multi-drug resistance and extended spectrum beta-lactamase- production in non-fermenting gram-negative bacilli in Ethiopia. Infect Dis Res Treat. 2019;12:1-7.
- 33. Beyene D, Bitew A, Fantew S, Mihret A, Evans M. Multidrug-resistant profile and prevalence of extended spectrum β-lactamase and carbapenemase- production in fermentative gram-negative bacilli recovered from patients and specimens referred to National Reference Laboratory, Addis Ababa, Ethiopia. PLoS One. 2019;14(9):e0222911. Crossref
- 34. Sonda T, Kumburu H, van Zwetselaar M, et al. Meta-analysis of proportion estimates of extended-spectrum-beta-lactamase-producing Enterobacteriaceae in East Africa hospitals. Antimicrob Resist Infect Control.2016;5:18. Crossref
- 35. Abrar S, Hussain S, Khan RA, Ul Ain N, Haider H, Riaz S. Prevalence of extended-spectrum-β-lactamase-producing Enterobacteriaceae: first systematic meta-analysis report from Pakistan. Antimicrob Resist Infect Control. 2018;7:26. Crossref
- 36. Tansarli GS, Poulikakos P, Kapaskelis A, Falagas ME. Proportion of extended-spectrum b-lactamase (ESBL)-producing isolates among Enterobacteriaceae in Africa: evaluation of the evidence—systematic review. J Antimicrob Chemother. 2014;69:1177-1184. Crossref
- 37. Mansouri F, Sheibani H, Masroor MJ, Afsharian M. Extended‐spectrum beta‐lactamase (ESBL)‐producing Enterobacteriaceae and urinary tract infections in pregnant/postpartum women: a systematic review and meta‐analysis. Int J Clin Pract. 2019;37(12). Crossref
- 38. Zhang J, Zheng B, Zhao L, et al. Nationwide high prevalence of CTX-M and an increase of CTX-M-55 in Escherichia coli isolated from patients with community-onset infections in Chinese county hospitals. BMC Infect Dis. 2014;14:659-663.
- 39. Obeng-Nkrumah N, Twum-Danso K, Krogfelt K, Newman MJ. High levels of extended-spectrum beta-lactamases in a major teaching hospital in Ghana: the need for regular monitoring and evaluation of antibiotic resistance. Am J Trop Med Hyg. 2013;89:960-964.
- 40. Magoué CL, Melin P, Gangoué-Piéboji J, Okomo Assoumou M-C, Boreux R, De Mol P. Prevalence and spread of extended-spectrum β-lactamase-producing Enterobacteriaceae in Ngaoundere, Cameroon. Clin Microbiol Infect. 2013;19:416-420.
- 41. Girlich D, Bouihat N, Poirel L, Benouda A, Nordmann P. High rate of faecal carriage of extended-spectrum β-lactamase and OXA-48 carbapenemase-producing Enterobacteriaceae at a university hospital in Morocco. Clin Microbiol Infect. 2014;20:350-354.
- 42. Hoban DJ, Lascols C, Nicolle LE, et al. Antimicrobial susceptibility of Enterobacteriaceae, including molecular characterization of extended-spectrum beta-lactamase-producing species, in urinary tract isolates from hospitalized patients in North America and Europe: results from the SMART study. Diagn Microbiol Infect Dis. Elsevier Inc. 2012;74(1):62-67. Crossref
- 43. Yang Q, Xu YC, Kiratisin P, Dowzicky MJ. Antimicrobial activity among gram-positive and gram-negative organisms collected from the Asia-Pacific region as part of the Tigecycline Evaluation and Surveillance Trial: comparison of 2015 results with previous years. Diagn Microbiol Infect Dis. 2017;89:314-323.
- 44. Hawser SP, Bouchillon SK, Hoban DJ, Badal RE, Hsueh PR, Paterson DL. Emergence of high levels of extended-spectrum-beta-lactamase-producing gram-negative bacilli in the Asia-Pacifc region: data from the Study for Monitoring Antimicrobial Resistance Trends (SMART) program, 2007. Antimicrob Agents Chemother. 2009;53:3280-3284.
- 45. Lu PL, Liu YC, Toh HS, et al. Epidemiology and antimicrobial susceptibility profiles of Gram-negative bacteria causing urinary tract infections in the Asia-Pacific region: 2009-2010 results from the Study for Monitoring Antimicrobial Resistance Trends (SMART). Int J Antimicrob Agents. 2012;40:S37-S43.
- 46. Lewis JM, Lester R, Garner P, Feasey NA. Gut mucosal colonisation with extended-spectrum beta-lactamase producing Enterobacteriaceae in sub-Saharan Africa: a systematic review and meta-analysis [version 2; peer- review: 2 approved]: Wellcome Open Res. 2020;4:160. Crossref
- 47. Muhie OA. Antibiotic use and resistance pattern in Ethiopia: systematic review and meta-analysis. Int J Microbiol. 2019, Article ID 2489063, 8 pages. Crossref
- 48. Pavez M, Troncoso C, Irma Osses I, et al. High prevalence of CTX-M-1 group in ESBL-producing Enterobacteriaceae infection in intensive care units in southern Chile. Braz J Infect Dis. 2019;23(2):102-110.
- 49. Moges F, Endris M, Belyhun Y, Worku W. Isolation and characterization of multiple drug resistance bacterial pathogens from waste water in hospital and non-hospital environments, Northwest Ethiopia. BMC Res Notes. 2014;7:215. Published 2014 Apr 5. Crossref
- 50. Rodríguez-Baño J, Gutiérrez-Gutiérrez B, Machuca I, Pascual A. Treatment of infections caused by extended-spectrum-beta-lactamase-, AmpC-, and carbapenemase-producing Enterobacteriaceae. Clin Microbiol Rev. 2018;31:e00079-17. Crossref
- 51. Harris P, Paterson D, Rogers B. Facing the challenge of multidrug-resistant gram-negative bacilli in Australia. MJA. 2015;202(5). Crossref
- 52. Seboxa T, Amogne W, Abebe W, et al. High mortality from blood stream infection in Addis Ababa, Ethiopia, is due to antimicrobial resistance. PLoS One. 2015;10(12):e0144944. Crossref
- 53. Flokas M, Karanika S, Alevizakos M, Mylonakis E. Prevalence of ESBL-producing Enterobacteriaceae in pediatric blood-stream infections: a systematic review and meta-analysis. PLoS One. 2017;12(1). Crossref