 |
Drug Target Insights 2020; 14: 26-33 DOI: 10.33393/dti.2020.2103 ORIGINAL RESEARCH ARTICLE |
 |
Targeting Streptococcus pneumoniae UDP-glucose pyrophosphorylase (UGPase): in vitro validation of a putative inhibitor
ABSTRACT
Background: Genome plasticity of Streptococcus pneumoniae is responsible for the reduced efficacy of various antibiotics and capsular polysaccharide-based vaccines. Therefore, targets independent of capsular types are sought to control the pneumococcal pathogenicity. UDP-glucose pyrophosphorylase (UGPase) is one such desired candidate being responsible for the synthesis of UDP-glucose, a sugar precursor in capsular biosynthesis and metabolic Leloir pathway. Being crucial to pneumococcal pathobiology, the effect of UGPase inhibition on virulence was evaluated in vitro.
Methods: A putative inhibitor, uridine diphosphate (UDP), was evaluated for effective inhibitory concentration in S. pneumoniae and A549 cells, its efficacy and toxicity. The effect of UDP on adherence and phagocytosis was measured in human respiratory epithelial (A549 and HEp-2) and macrophage (THP1 and J774.A.1) cell lines respectively.
Results: A differential effective inhibitory concentration of UDP for UGPase inhibition was observed in S. pneumoniae and A549 cells, that is, 5 and 100 µM respectively. UDP treatments lowered percent cytotoxicity in pneumococcal-infected monolayers and didn’t exert adverse effects on viabilities. S. pneumoniae adherence to host cells decreased significantly with UDP treatments. UDP induced the secretion of interleukin (IL)-1β, tumor necrosis factor (TNF)-α, IL-6, and IL-8 and increased pneumococcal phagocytosis.
Conclusion: Our study shows UDP-mediated decrease in the virulence of S. pneumoniae and demonstrates UDP as an effective inhibitor of pneumococcal UGPase.
Keywords: Capsule, galU, Inhibitor, Pneumococcus, UDP, Virulence
Received: February 26, 2020
Accepted: July 30, 2020
Published online: October 7, 2020
© 2020 The Authors. This article is published by AboutScience and licensed under Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0). Any commercial use is not permitted and is subject to Publisher’s permissions. Full information is available at www.aboutscience.eu
Introduction
Streptococcus pneumoniae (pneumococcus) is an important healthcare-associated pathogen being responsible for various invasive pneumococcal diseases (IPDs), for example, bacteremia, pneumonia, septicemia, and meningitis, and non-IPDs, for example, acute otitis media and sinusitis (1). Pneumococcal infections are associated with significant morbidity and mortality globally, specifically in children younger than 5 years, elderly adults, and individuals with immune deficiencies. Global disease burden and mortality estimates have documented 341,029 deaths in children aged below 5 years; 494,340 deaths in the elderly (aged above 70 years), and total 1,189,937 deaths among all ages due to pneumococcal infections (2,3,4). This huge burden of pneumococcal diseases is fueled by the rise of new capsular serotypes and spread of antimicrobial-resistant clones (5). Introduction of the capsular polysaccharide (CPS)-based vaccines and pneumococcal conjugate vaccines had provided a little relief, but they still remain unsatisfactory due to limited serotype protection and replacement, restricted efficacy of vaccine at the mucosal surface, implementation issues, and high cost (6,7). Hence, formulation of alternative preventive measures independent of serotypes is inevitable for the better management of pneumococcal diseases.
Pneumococcal strains have been shown to synthesize a distinct serotype-specific polysaccharide capsule capable of dodging host immune components (8,9). The loss of capsule has been associated with rapid phagocytosis (10,11). Genetic dissection of the cps/cap gene cluster encoding for CPS had strongly advocated the involvement of other unlinked genes that are dispersed in chromosome in CPS synthesis (12,13). One of the genes, galU, encodes for enzyme uridine diphosphate (UDP)-glucose pyrophosphorylase (UGPase), and its role in the virulence of various gram-negatives and gram-positives is well studied (14,15,16,17,18,19). UGPase catalyzes the synthesis of UDP-glucose (UDP-glc), which is an important glycosyl donor for the modification and interconversion of sugars in metabolic and capsular pathways (17,19,20). The galU mutants of pneumococcus have been shown to be acapsular despite the presence of functional cps genes (14,19).
UGPase has been considered as a suitable target to control the pneumococcal virulence (17,21) as galU has been reported in all pneumococcal strains regardless of capsular types (14). Also, mammalian UGPases have been evolutionarily unrelated to their prokaryotic counterparts, suggesting that its putative inhibitors would not be inimical to the host (17,21,22). The effects of UGPase inhibition in the pathogenicity of pneumococcus have not been elucidated so far despite the availability of evidence of gene mutation studies (19). A study has screened few chemical inhibitors for the inhibition of UGPase activity in a calorimetric assay from purified extracts (23), but no study is available on inhibitor-mediated host-pathogen interaction. Therefore, for the selection of UGPase inhibitor, we modeled and analyzed the structure of S. pneumoniae UGPase using I-TASSER (Iterative-Threading ASSEmbly Refinement) server (http://zhang.bioinformatics.ku.edu/I-TASSER) to delineate its tertiary structure, active site residues, and properties. In this study, we have studied the effect of UGPase inhibition on the virulence of S. pneumoniae during host-pathogen interactions and validated a putative inhibitor of pneumococcal UGPase for its inhibition potentials and efficacy in vitro.
Materials and methods
This study was approved by the Institute Ethics Committee (IEC Memo no. 87770-PG11-ITRG/11384). Respiratory epithelial (A549 and HEp-2) and macrophage (J774.A.1 and THP1) cell lines were procured from the cell repository of National Centre for Cell Sciences (NCCS), Pune, India. S. pneumoniae reference strains D39 (NCTC 7466) and MTCC 655 (NCTC 7465) were procured from Microbial Type Culture Collection (MTCC), IMTECH, Chandigarh, India. A clinical blood isolate of serotype 19F was also used in the study as its occurrence has been reported frequently in IPDs. Clinical strain was isolated from the blood sample of a patient suffering from lower respiratory tract infection of S. pneumoniae only.
Cell culture
A549 and THP1 cell lines were cultured and maintained in RPMI 1640, while HEp-2 and J774.A.1 cell lines were cultured in Minimal Essential Medium (MEM) and Dulbecco’s Modified Eagle’s Medium (DMEM), respectively, at 37°C with 5% CO2. All the media were supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS), 0.15% (v/v) sodium bicarbonate, streptomycin (100 µg/mL), and penicillin (100 U/mL). Antibiotic-free media were prepared for experiments involving bacterial infection.
UDP as UGPase inhibitor
Effective concentration of UDP as inhibitor of UGPase was evaluated by UGPase assay (17) in both S. pneumoniae and A549 cells. Log phase culture of S. pneumoniae and confluent cell monolayers were treated with UDP at different concentrations for 1 hour at 37°C and 5% CO2. A549 cells in phosphate-buffered saline (PBS) were lysed with three cycles of temperature-shock (5 minutes each). Bacterial pellets were sonicated (3 cycles; pulse 10 seconds ON, 30 seconds OFF) in ice bath. UGPase activity was assessed in total 1 mL reaction mixture with freshly prepared extracts (100 μL) at 25°C. Reaction mixture constituted 50 mM Tris-HCl buffer (pH 7.5), 16 mM MgCl2, 0.6 mM β-nicotinamide adenine dinucleotide phosphate (NADP), 0.6 mM UDP-glucose, UDP-glucose dehydrogenase (0.7 U), and phosphoglucomutase (0.07 U). Reaction was initiated with the addition of sodium pyrophosphate (1.7 mM) and NADPH formation was determined by measuring the increase in absorbance at 340 nm. Blank consisted of reaction mixture without cell or bacterial extract.
Inhibitor toxicity to host cells
A549 cell monolayers were treated with different concentrations of UDP for 1 hour at 37°C and untreated cells were taken as reference. The effect of UDP on viability of cells was evaluated using MTT assay (24). Briefly, MTT solution (2.5 mg/mL; Sigma) was added to monolayers and incubated for 4 hours. Media containing MTT was discarded and dimethyl sulfoxide (DMSO, 200 µL) was added. Absorbance was recorded at 590 nm in microplate reader (Bio-Rad 680).
Expression of UGPase in host cells
A549 and HEp-2 monolayers were treated with UDP at effective inhibiting dose for 1 hour at 37°C, 5% CO2 and untreated cells were taken as control. Total protein was extracted from cell pellet by sonication (3 minutes: pulse 30 seconds ON, 30 seconds OFF) in ice-chilled PBS and was quantitated. Samples were electrophoresed in 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and separated proteins were transferred onto polyvinylidene difluoride (PVDF) membrane. Membranes were processed and incubated with anti-UGP2 antibody (1:2,000, Abcam) for 3 hours at 37°C followed by treatment with horseradish peroxidase (HRP)-conjugated secondary antibody (1:20,000) for 2 hours at 37°C. Pierce® ECL western blotting peroxidase substrate (ThermoScientific) was used for developing protein bands, which were quantitated with FluorChem M ProteinSimple (Bio-Techne). GAPDH as endogenous control was simultaneously blotted and analyzed. Expression of UGPase was normalized with the value of GAPDH in each sample.
Adherence assay
Efficacy of UDP to inhibit the adherence of S. pneumoniae during infection with A549 and HEp-2 monolayers was checked by flow cytometer (BD Biosciences) (25). Log phase cultures of pneumococcus were labeled with fluorescein isothiocyanate (FITC, 1.0 μg/μL) and infected onto monolayers at 100:1 multiplicity of infection (MOI: bacteria/cell) in the presence and absence of inhibitor at effective dose. Monolayers were incubated at 37°C at 5% CO2 for 2 hours followed by trypsinization (0.05%) and fixation with ice-cold paraformaldehyde (2%). Samples were analyzed using FACScan flow cytometer using CellQuest 3.3 software (BD Biosciences). Mean fluorescence intensity (MFI) values were obtained from control samples (unlabeled host cells and bacteria). Adherence was calculated from the MFI of host cell with adherent pneumococci divided by MFI of labeled pneumococci. Effectiveness of inhibitor was evaluated from the adherence potential of S. pneumoniae to monolayers in the presence of inhibitor.
Phagocytosis assay
FITC-labeled pneumococci were loaded (MOI, 100:1) onto macrophage cells (THP1 and J774.A.1) in the presence and absence of UDP for 1 hour at 37°C in 5% CO2. Cells were fixed with cold 2% paraformaldehyde. Briefly, cells were collected, centrifuged, washed as required, and were suspended in ice-cold PBS. Trypan blue (0.2%) was added to quench fluorescence of extracellularly attached pneumococci, just before analysis in flow cytometer (BD BioSciences). MFI of FITC-positive cells was calculated to assess the phagocytic activity in comparison to controls (unlabeled cells and bacteria).
For opsonophagocytic assay (OPA), bacterial cells (106 cfu/mL) were labeled with FITC (1 μg/μL) after inactivation at 95°C for 5 minutes. Nonviability was confirmed by overnight incubation of blood agar plates streaked with bacterial suspension (100 μL). Labeled bacteria were incubated with pooled human sera (10% of 1:2 diluted in PBS) for 30 minutes at 37°C, 150 rpm. Bacteria were further inoculated onto THP1 and J774.A.1 cells at MOI 100:1 for 1 hour at 37°C and 5% CO2. Control tubes contained PBS or heat-inactivated (56°C for 45 minutes) pooled sera. The cells were obtained, washed, centrifuged, and fixed with cold 2% paraformaldehyde and analyzed in a flow cytometer (26).
Expression of postinfection cytokines
A549 monolayers were incubated with effective dose of UDP for 1 hour at 37°C, 5% CO2. In another set, monolayers were treated with 10 μM MRS2578 for 30 minutes before incubation with UDP (27). Total ribonucleic acid (RNA) was extracted from trypsinized cell pellets using TRIzol reagent followed by complementary deoxyribonucleic acid (cDNA) synthesis using messenger RNA RevertAid™ First Strand cDNA Synthesis Kit (Thermo Scientific). Expression of various cytokines (interleukin [IL]-6, IL-1β, IL-8, tumor necrosis factor [TNF]-α) was analyzed using real-time polymerase chain reaction (PCR; Roche Lightcycler® 480) in reactions (10 μL) containing 50 ng cDNA, 0.5 μM primer, 1X SYBR mix, and deionized water. Gene-specific primers were designed using Primer3Tool and PCRs were run in primer-specific cycling conditions and with appropriate negative controls for each reaction.
Statistical analysis
Each experiment was performed at least three times in duplicate or triplicate sets. GraphPad Prism 6.0 was used for statistical calculations. Student’s t-test, or Mann-Whitney U-test or analysis of variance (ANOVA) was applied as appropriate. A p value less than 0.05 was considered to represent a significant association.
Results
Putative inhibitor of UGPase
The structure of S. pneumoniae UGPase was analyzed in silico using I-TASSER server (http://zhang.bioinformatics.ku.edu/I-TASSER; PhD Thesis). The tertiary structure, active site residues, and its substrate-binding properties were analyzed. The local conformational changes induced near the active site in response to binding of its natural substrate were analyzed. Using information of the active site pocket of UGPase and literature-based evidence (11,14,17,21,22,23), UDP was selected as a probable inhibitor of S. pneumoniae UGPase. In silico analysis showed that UDP binding did not result in alteration in local conformation of enzymatic active site. The effective inhibitory concentrations of UDP, its extent of inhibition, and probable toxicity to host cell lines were studied. The pneumococcal virulence and clearance of infection in response to UDP were explored in vitro.
Effect of UDP on UGPase activity
UGPase activity was evaluated at different UDP concentrations (0.05-100 μM) in A549 cells and S. pneumoniae D39 strain. Concentration-dependent dose response curve showed that UDP decreased UGPase activity in host A549 cells (Fig. 1A) as well as in pneumococcus (Fig. 1B). UDP-treated A549 cells showed a declined UGPase activity in comparison to untreated (0.0 μM) control cells (Fig. 1A). UGPase activity was not significantly decreased in the cells treated with 0.1-3 μM UDP. However, activity was significantly decreased at 5 μM (p = 0.009) and 100 μM (p = 0.011) UDP treatments. The inhibition of UGPase activity in A549 cells was 2.8 fold higher at 100 μM UDP treatment in comparison to 5 μM treatment.
In S. pneumoniae D39 strain, a significant decrease in UGPase activity was observed at 2 μM UDP (p = 0.02) treatment (Fig. 1B). At 5 μM UDP, threefold inhibition (p = 0.01) was observed in UGPase activity in comparison to 100 μM. Further, effect of UDP (at 5 and 100 μM) on UGPase activity was evaluated in invasive S. pneumoniae isolate (retrieved from blood sample), which showed a decrease in activity at 5 μM UDP (p = 0.015) (Fig. 1C). Comparative analysis of UDP treatment on blood isolate and reference D39 strain showed that 5 μM UDP treatments for 1 hour were effective to reduce the bacterial UGPase activity (Fig. 1C) by half than in its host counterpart (Fig. 1A).
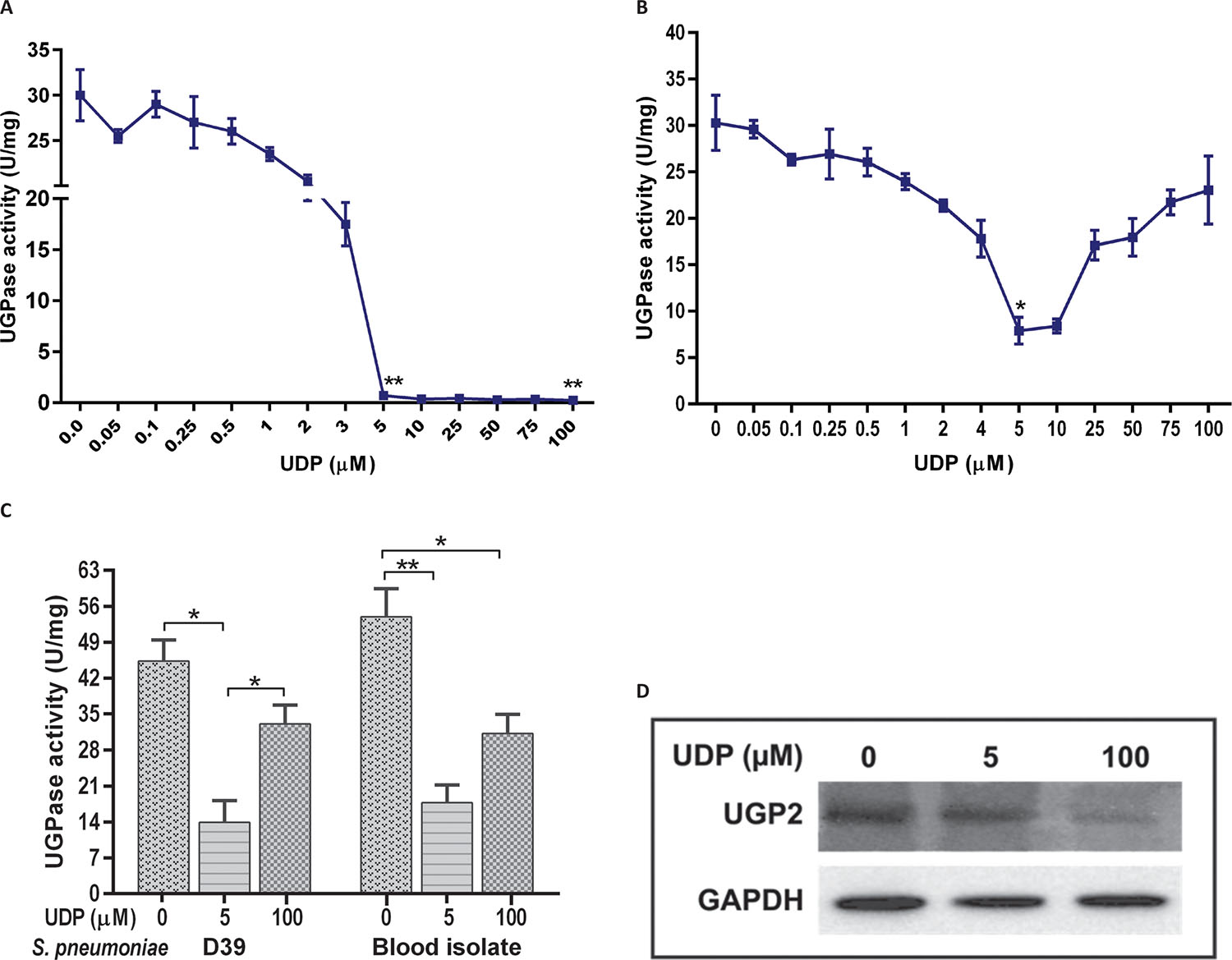
Further analysis of UGPase expression in A549 cell lysate using human anti-UGP2 antibody reconfirmed the significant decrease (p = 0.03) at 100 μM UDP treatment in comparison to 5 μM treated and control (untreated) cells (Fig. 1D). However, we could not analyze the UGPase expression in S. pneumoniae due to unavailability of antibody against bacterial enzymes.
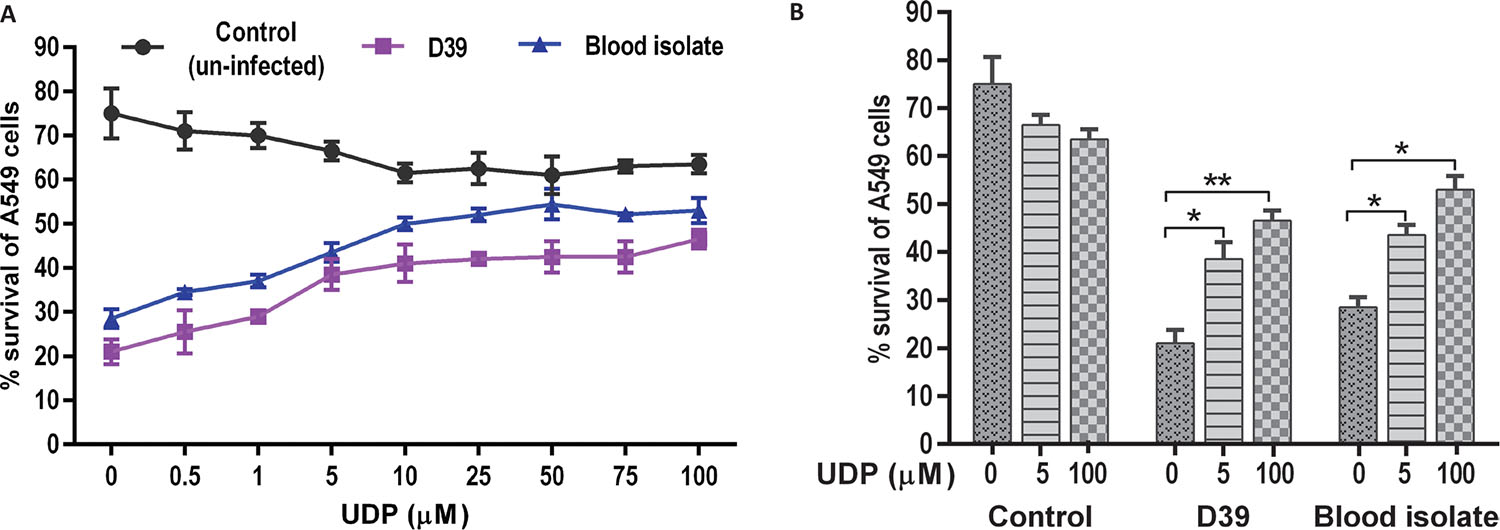
UDP treatment and A549 cell survival
The effect of UDP on A549 cell viability was evaluated in a dose-dependent manner in the absence (control) and presence of S. pneumoniae infection (Fig. 2). Uninfected (control) cells treated with UDP (0.5-100 μM) showed negligible changes in the percentage cell viability. However, A549 cells infected with pneumococci (D39 and blood isolate) showed a significant decrease in percent viability in the absence of UDP treatment (p = 0.001). Notably, the viability of infected A549 cells was elevated at higher UDP treatments (>10 μM) in a dose-dependent manner (Fig. 2A). Further, a comparative analysis of UDP treatments (0, 5, 100 μM) to A549 cells showed significantly improved viability at 100 μM treatment in D39 (p = 0.009) and blood isolate (p = 0.04) infected cells, while no change in cell viability was observed in uninfected (control) cells (Fig. 2B). Thus, UDP seems safer to explore for its potential as it did not exert any cytotoxic effect on A549 cells (Fig. 2), while it improved the percentage cell viability in the presence of bacterial infection.
Further, effect of UGPase inhibition on adherence and phagocytosis of S. pneumoniae (blood isolate and standard strains MTCC 655; a highly capsular strain and D39) was evaluated at effective 5 μM UDP concentration (Fig. 1).
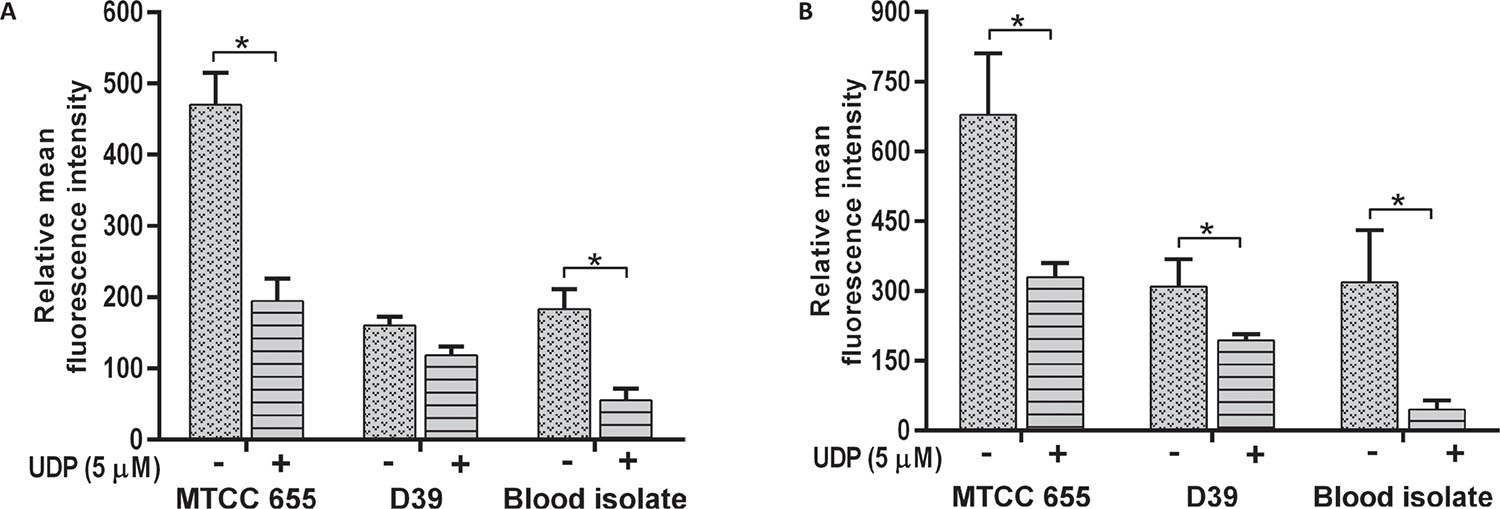
Effect of UDP on S. pneumoniae adherence
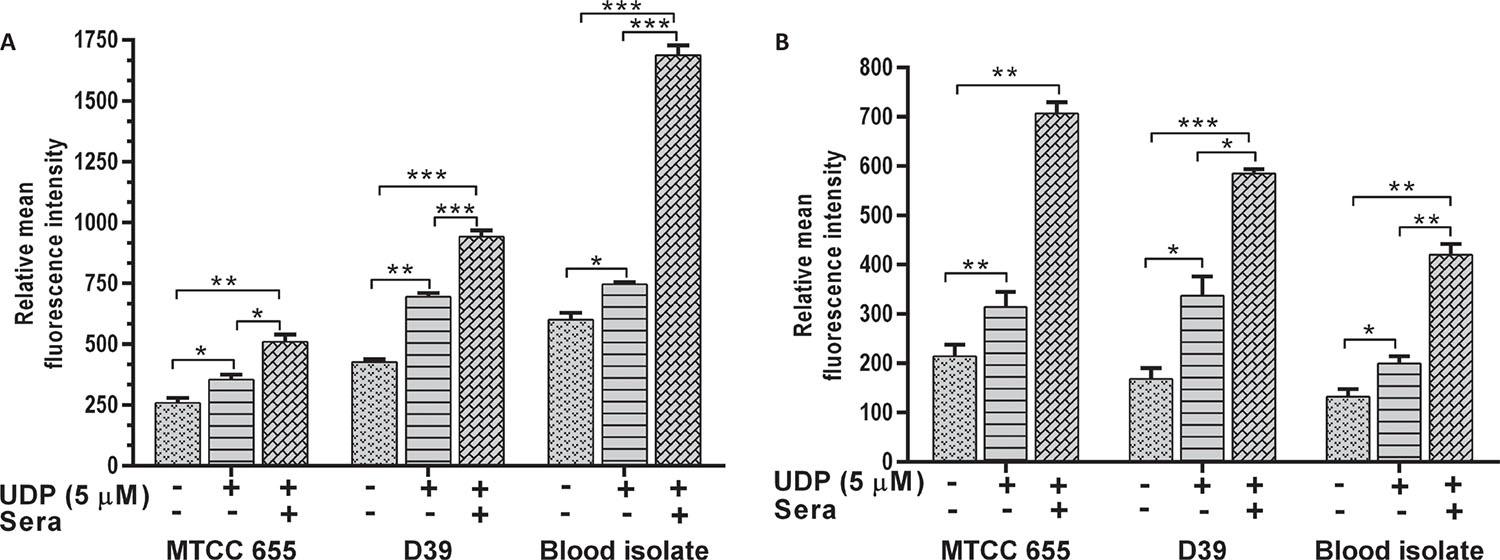
All S. pneumoniae strains showed a decrease in adherence to A549 (p = 0.018) and HEp-2 cells (p = 0.001) in the presence of 5 μM UDP (Fig. 3). Adherence of MTCC 655 and blood isolate was significantly (p<0.05) lowered in both the cell lines (Fig. 3A and B), while D39 showed a significant (p = 0.031) decrease in HEp-2 cells only (Fig. 3B). Adherence of blood isolate to A549 (p = 0.032) and HEp-2 (p = 0.014) cells declined by three- to fivefold.
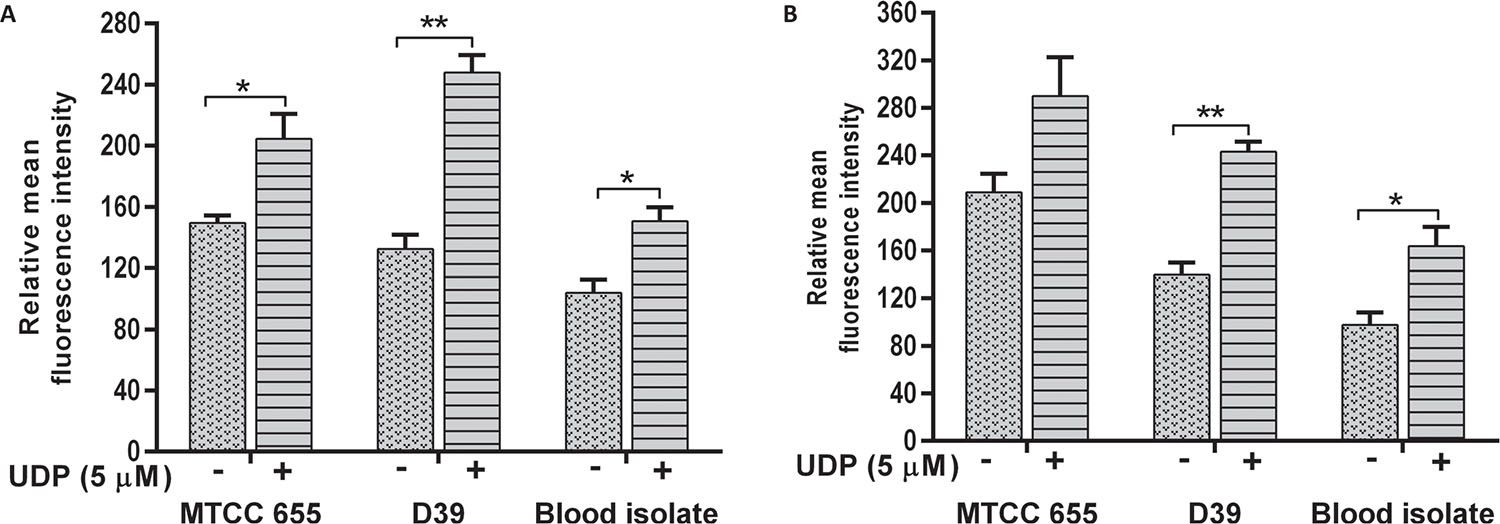
Effect of UDP on phagocytosis of S. pneumoniae
The phagocytosis of S. pneumoniae was increased in the presence of UDP in both J774.A.1 (p = 0.01) and THP1 (p = 0.022) cells (Fig. 4A and B respectively). Phagocytosis of D39 strain was significantly higher than MTCC 655 strain (p = 0.033) in both the cell lines, which could be attributed to heavy encapsulation in the latter. UDP treatment also showed an increase in phagocytosis of blood isolate by J774.A.1 (p = 0.034) and THP1 cells (p = 0.041). Overall, UDP-dependent fold change in phagocytosis ranged from 1.4 to 1.9 for pneumococcal strains. It is well known that host immune cells use complement factors for the clearance of S. pneumoniae, and OPA of heat-inactivated S. pneumoniae was found to be enhanced in the presence of UDP in both the cell lines (Fig. 5A and B). Interestingly, J774.A.1 cells (Fig. 5A) showed marked increase in the OPA of blood isolate than MTCC 655 and D39 strains. Phagocytosis of MTCC 655 and D39 strains was higher in THP1 cells (Fig. 5B) in the presence of UDP (5 μM) and sera.
Real-time PCR analysis of cytokines
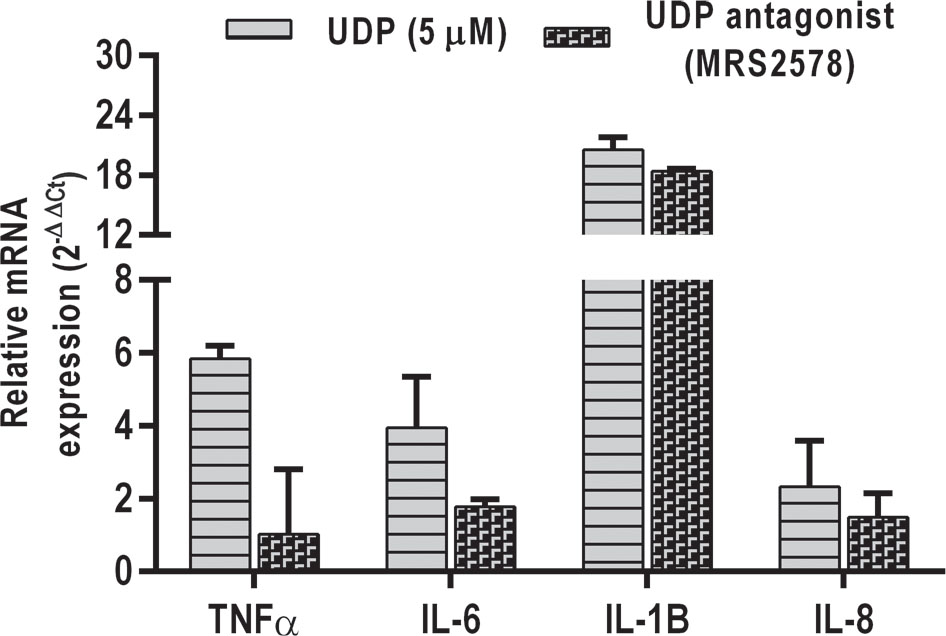
An attempt was made to evaluate the cytokine response in A549 cells in response to UDP treatment as previous studies have shown the activation of inflammatory pathway upon UDP stimulation from monocytes and microglial cells (28,29). Interestingly, UDP treatment (5 μM) induced the secretion of IL-1β, TNF-α, IL-6, and IL-8 from the A549 alveolar epithelial cells (Fig. 6). An UDP antagonist MRS2578 (Sigma) effectively suppressed the expression of these cytokines in A549 cells. In agreement with these results, UDP is shown to bind its purinergic receptor P2Y6 on various cells and stimulate signaling cascade (30,31).
Conclusions
The paradox of infections caused by S. pneumoniae lies in the availability of preventive measures such as antibiotics and vaccines, but not effective prevention from the pneumococcal diseases, especially in low- to middle-income settings. The major reason for this paradox is the genome plasticity of pneumococcus and diverse nature of cps loci, which has precluded the absolute success of vaccines (11,12,32). Evidently, various virulence factors of pneumococci had been explored to understand its pathobiology and with broader prospect to yield potential targets for preventive interventions (8,9,10,11,21). Elucidation of cps loci from different pneumococcal serotypes has established a crucial role of galU-encoded UGPase in capsule formation (14,17,18). UGPase-catalyzed reaction provides UDP-glucose (glc), a key sugar precursor to fulfill various capsular (CPS biosynthesis pathway) and metabolic (Leloir pathway) needs of pneumococcus (12,13,14,18,20,36,37,38). In this study, we explored the potential of UGPase in modulating S. pneumoniae pathogenicity by using UDP as an UGPase inhibitor.
UDP proved to be an effective inhibitor of pneumococcal UGPase and mediated a significant reduction in the in vitro virulence of pneumococci. The differential concentration-based enzyme activity inhibition maxima suggests the feasibility of selective inhibition of pneumococcal UGPase (5 μM UDP) without inimical effect on host. The decline in pneumococcal adherence to host cell lines also accentuates the efficacy of UDP as an inhibitor. A study by Zavala et al used nucleoside analogs, namely abacavir, decitabine, stavudine, and zidovudine, to show 42%-58% inhibition of UGPase, but did not elaborate on the use of high concentration (7.5 mM) of inhibitors, their toxic effects, and safety as antipneumococcal drug (23). Our study establishes the noncytotoxicity of UDP to host cells and boosted uptake of S. pneumoniae upon UDP stimulation by macrophage cell lines. Our findings are supported by previous studies on the clearance of Escherichia coli from peritonitis mouse model in response to direct UDP injection (33) and increased host defenses upon UDP binding to P2Y6 receptor (3435,36). The crucial role of UDP as inflammatory inducer in brain injuries has been reported in microglia and astrocytes (28). Likewise, we have also found the activation of proinflammatory cytokines in alveolar epithelial cells in response to pneumococcal infection. The stimulation of cytokines by UDP emphasizes its protective functional nature, which might enhance its efficacy as a potent inhibitor.
The present study provides evidence to the UDP-mediated reduction in the virulence of S. pneumoniae. Being one of the intercellular messengers, UDP acts as a protective signaling molecule and activates P2Y6 receptors that are known to result in wide range of physiologic responses, such as induction of cytokines, chemokines, phagocytosis, and increase in concentration of extracellular nucleotides (33,35,37,38). The evidence of UDP meddling with host-pathogen interactions has opened the vast horizon to investigate its underlying mechanisms, which would definitely shed light on its potential utility in restraining the pneumococcal virulence. The present work is a little step forward in search of potential S. pneumoniae inhibitor independent of serotypes, though the inhibitory effect of UDP should be evaluated in vivo to ascertain its efficacy. Our study has been limited in evaluating the cytokine response and UDP-mediated behavior of P2Y6 receptors, which further need a detailed investigation. Mechanistic understanding of UDP-mediated UGPase inhibition and its in vivo validation would be crucial for translating the outcome of this study.
Author contributions
Monica Sharma performed the experiments, analyzed the data, and wrote the manuscript. Swati Sharma helped in manuscript editing. Anuradha Chakraborti and Monica Sharma designed the study. Pallab Ray provided the clinical isolates for the study. Anuradha Chakraborti and Pallab Ray reviewed the study and manuscript.
Disclosures
Financial support: The authors are grateful to the Indian Council of Medical Research (ICMR), New Delhi, India for financial support and providing Senior Research Fellowship (File no. 80/768/2012-ECD-1) to Ms. Monica Sharma and funding the project. Swati Sharma received research fellowship from Council of Scientific and Industrial Research (CSIR) New Delhi, India. Authors disclose no other external funding sources.
Conflict of interest: The authors declare no conflict of interest.
References
- 1. Musher DM. Streptococcus pneumoniae. In: Mandell GL, Bennett JE, Dolin R, eds. Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. 7th ed. Philadelphia, PA: Churchill Livingstone Elsevier 2010; 2623-2642.
- 2. Troeger C, Blacker B, Khalil IA, et al., SRM GBD 2016 Lower Respiratory Infections Collaborators. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of lower respiratory infections in 195 countries, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect Dis. 2018;18:1191-1210.
- 3. Wahl B, O’Brien KL, Greenbaum A, et al. Burden of Streptococcus pneumoniae and Haemophilus influenzae type b disease in children in the era of conjugate vaccines: global, regional, and national estimates for 2000-15. Lancet Glob Health. 2018;6:e744-e757.
- 4. Wahl B, Sharan A, Deloria Knoll M, et al. National, regional, and state-level burden of Streptococcus pneumoniae and Haemophilus influenzae type b disease in children in India: modeled estimates for 2000-15. Lancet Glob Health. 2019;7:e735-e747.
- 5. Kim SH, Song JH, Chung DR, et al. Changing trends in antimicrobial resistance and serotypes of Streptococcus pneumoniae isolates in Asian countries: an Asian Network for Surveillance of Resistant Pathogens (ANSORP) study. Antimicrob Agents Chemother. 2012;56:1418-1426.
- 6. Azarian T, Grant LR, Arnold BJ, et al. The impact of serotype-specific vaccination on phylodynamic parameters of Streptococcus pneumoniae and the pneumococcal pan-genome. PLoS Pathog. 2018;14:e1006966.
- 7. McLaughlin JM, Jiang Q, Isturiz RE, et al. Effectiveness of 13-valent pneumococcal conjugate vaccine against hospitalization for community-acquired pneumonia in older US adults: a test-negative design. Clin Infect Dis. 2018;67:1498-1506.
- 8. Mitchell AM, Mitchell TJ. Streptococcus pneumoniae: virulence factors and variation. Clin Microbiol Infect. 2010;16:411-418.
- 9. Brooks LRK, Mias GI. Streptococcus pneumoniae’s virulence and host immunity: aging, diagnostics, and prevention. Front Immunol. 2018;9:1366.
- 10. Magee AD, Yother J. Requirement for capsule in colonization by Streptococcus pneumoniae. Infect Immun. 2001;69:3755-3761.
- 11. Shainheit MG, Mulé M, Camilli A. The core promoter of the capsule operon of Streptococcus pneumoniae is necessary for colonization and invasive disease. Infect Immun. 2014;82:694-705.
- 12. García E, Llull D, López R. Functional organization of the gene cluster involved in the synthesis of the pneumococcal capsule. Int Microbiol. 1999;2:169-176.
- 13. García E, Llull D, Muñoz R, Mollerach M, López R. Current trends in capsular polysaccharide biosynthesis of Streptococcus pneumoniae. Res Microbiol. 2000;151:429-435.
- 14. Mollerach M, López R, García E. Characterization of the galU gene of Streptococcus pneumoniae encoding a uridine diphosphoglucose pyrophosphorylase: a gene essential for capsular polysaccharide biosynthesis. J Exp Med. 1998;188:2047-2056.
- 15. Chang HY, Lee JH, Deng WL, Fu TF, Peng HL. Virulence and outer membrane properties of a galU mutant of Klebsiella pneumoniae CG43. Microb Pathog. 1996;20:255-261.
- 16. Priebe GP, Dean CR, Zaidi T, et al. The galU gene of Pseudomonas aeruginosa is required for corneal infection and efficient systemic spread following pneumonia but not for infection confined to the lung. Infect Immun. 2004;72:4224-4232.
- 17. Bonofiglio L, García E, Mollerach M. Biochemical characterization of the pneumococcal glucose 1-phosphate uridylyltransferase (GalU) essential for capsule biosynthesis. Curr Microbiol. 2005;51:217-221.
- 18. Bonofiglio L, García E, Mollerach M. The galU gene expression in Streptococcus pneumoniae. FEMS Microbiol Lett. 2012;332:47-53.
- 19. Cools F, Torfs E, Vanhoutte B, et al. Streptococcus pneumoniae galU gene mutation has a direct effect on biofilm growth, adherence and phagocytosis in vitro and pathogenicity in vivo. Pathog Dis. 2018;76.
- 20. Frey PA. The Leloir pathway: a mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose. FASEB J. 1996;10:461-470.
- 21. Alvaro Berbis M, Maria Sanchez-Puelles J, Javier Canada F, Jimenez-Barbero J. Structure and function of prokaryotic UDP-glucose pyrophosphorylase, a drug target candidate. Curr Med Chem. 2015;22:1687-1697.
- 22. Mollerach M, García E. The galU gene of Streptococcus pneumoniae that codes for a UDP-glucose pyrophosphorylase is highly polymorphic and suitable for molecular typing and phylogenetic studies. Gene. 2000;260:77-86.
- 23. Zavala A, Kovacec V, Levín G, et al. Screening assay for inhibitors of a recombinant Streptococcus pneumoniae UDP-glucose pyrophosphorylase. J Enzyme Inhib Med Chem. 2017;32:203-207.
- 24. Twentyman PR, Luscombe M. A study of some variables in a tetrazolium dye (MTT) based assay for cell growth and chemosensitivity. Br J Cancer. 1987;56:279-285.
- 25. Hara-Kaonga B, Pistole TG. A dual fluorescence flow cytometric analysis of bacterial adherence to mammalian host cells. J Microbiol Methods. 2007;69:37-43.
- 26. Hu BT, Yu X, Jones TR, et al. Approach to validating an opsonophagocytic assay for Streptococcus pneumoniae. Clin Diagn Lab Immunol. 2005;12:287-295.
- 27. Kim B, Jeong HK, Kim JH, Lee SY, Jou I, Joe EH. Uridine 5′-diphosphate induces chemokine expression in microglia and astrocytes through activation of the P2Y6 receptor. J Immunol. 2011;186:3701-3709.
- 28. Cox MA, Gomes B, Palmer K, et al. The pyrimidinergic P2Y6 receptor mediates a novel release of proinflammatory cytokines and chemokines in monocytic cells stimulated with UDP. Biochem Biophys Res Commun. 2005;330:467-473.
- 29. Grbic DM, Degagné É, Larrivée JF, et al. P2Y6 receptor contributes to neutrophil recruitment to inflamed intestinal mucosa by increasing CXC chemokine ligand 8 expression in an AP-1-dependent manner in epithelial cells. Inflamm Bowel Dis. 2012;18:1456-1469.
- 30. Ginsburg-Shmuel T, Haas M, Grbic D, et al. UDP made a highly promising stable, potent, and selective P2Y6-receptor agonist upon introduction of a boranophosphate moiety. Bioorg Med Chem. 2012;20:5483-5495.
- 31. Hao Y, Liang JF, Chow W, Cheung W, Ko W. P2Y6 receptor-mediated proinflammatory signaling in human bronchial epithelia. PLoS ONE. 2014;9:e106235.
- 32. Subramanian K, Henriques-Normark B, Normark S. Emerging concepts in the pathogenesis of the Streptococcus pneumoniae: from nasopharyngeal colonizer to intracellular pathogen. Cell Microbiol. 2019;2:e13077.
- 33. Zhang Z, Wang Z, Ren H, et al. P2Y6 agonist uridine 5′-diphosphate promotes host defense against bacterial infection via monocyte chemoattractant protein-1–mediated monocytes/macrophages recruitment. J Immunol. 2011;186:5376-5387.
- 34. Idzko M, Ferrari D, Eltzschig HK. Nucleotide signalling during inflammation. Nature. 2014;509:310-317.
- 35. Koizumi S, Shigemoto-Mogami Y, Nasu-Tada K, et al. UDP acting at P2Y6 receptors is a mediator of microglial phagocytosis. Nature. 2007;446:1091-1095.
- 36. Inoue K, Koizumi S, Kataoka A, Tozaki-Saitoh H, Tsuda M. P2Y(6)-evoked microglial phagocytosis. Int Rev Neurobiol. 2009;85:159-163.
- 37. Neher JJ, Neniskyte U, Hornik T, Brown GC. Inhibition of UDP/P2Y6 purinergic signaling prevents phagocytosis of viable neurons by activated microglia in vitro and in vivo. Glia. 2014;62:1463-1475.
- 38. Zierhut M, Dyckhoff S, Masouris I, et al. Role of purinergic signaling in experimental pneumococcal meningitis. Sci Rep. 2017;7:44625.