 |
Drug Target Insights 2020; 14: 1-11 DOI: 10.33393/dti.2020.1548 ORIGINAL RESEARCH ARTICLE |
 |
Identification of the possible therapeutic targets in the insulin-like growth factor 1 receptor pathway in a cohort of Egyptian hepatocellular carcinoma complicating chronic hepatitis C type 4
ABSTRACT
Background: Molecular targeted drugs are the first line of treatment of advanced hepatocellular carcinoma (HCC) due to its chemo- and radioresistant nature. HCC has several well-documented etiologic factors that drive hepatocarcinogenesis through different molecular pathways. Currently, hepatitis C virus (HCV) is a leading cause of HCC. Therefore, we included a unified cohort of HCV genotype 4-related HCCs to study the expression levels of genes involved in the insulin-like growth factor 1 receptor (IGF1R) pathway, which is known to be involved in all aspects of cancer growth and progression.
Aim: Determine the gene expression patterns of IGF1R pathway genes in a cohort of Egyptian HCV-related HCCs. Correlate them with different patient/tumor characteristics. Determine the activity status of involved pathways.
Methods: Total ribonucleic acid (RNA) was extracted from 32 formalin-fixed paraffin-embedded tissues of human HCV-related HCCs and 6 healthy liver donors as controls. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) using RT2 Profiler PCR Array for Human Insulin Signaling Pathway was done to determine significantly up- and downregulated genes with identification of most frequently coregulated genes, followed by correlation of gene expression with different patient/tumor characteristics. Finally, canonical pathway analysis was performed using the Ingenuity Pathway Analysis software.
Results: Six genes – AEBP1, AKT2, C-FOS, PIK3R1, PRKCI, SHC1 – were significantly overexpressed. Thirteen genes – ADRB3, CEBPA, DUSP14, ERCC1, FRS3, IGF2, INS, IRS1, JUN, MTOR, PIK3R2, PPP1CA, RPS6KA1 – were significantly underexpressed. Several differentially expressed genes were related to different tumor/patient characteristics. Nitric oxide and reactive oxygen species production pathway was significantly activated in the present cohort, while the growth hormone signaling pathway was inactive.
Conclusions: The gene expression patterns identified in this study may serve as possible therapeutic targets in HCV-related HCCs. The most frequently coregulated genes may serve to guide combined molecular targeted therapies. The IGF1R pathway showed evidence of inactivity in the present cohort of HCV-related HCCs, so targeting this pathway in therapy may not be effective.
Keywords: Gene expression, Hepatitis C virus, Hepatocellular carcinoma, Molecular therapeutic targets
Received: January 8, 2020
Accepted: January 20, 2020
Published online: April 8, 2020
© 2020 The Authors. This article is published by AboutScience and licensed under Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0). Any commercial use is not permitted and is subject to Publisher’s permissions. Full information is available at www.aboutscience.eu.
Introduction
Hepatocellular carcinoma (HCC) has become the fifth most common cancer in men worldwide and the ninth in women and the third leading cause of cancer-related deaths. Distinct geographical variations in the incidence exist. In Egypt, HCC has the highest age-standardized incidence and mortality rates (ASR) from cancer in males and ranks second in females (1). Most of the burden is in developing countries where 83% of cases (and deaths) occur (2). HCC has several well-documented etiologic factors that drive hepatocarcinogenesis through different molecular pathways. Currently, hepatitis C virus (HCV) is a leading cause of HCC. The multiplicity of etiologies of HCC and the existence of substantial molecular heterogeneity might limit the benefits of targeted approaches to HCC treatment. Therefore, therapeutic vulnerabilities should be studied in homogeneous groups of HCCs sharing similar drivers of the disease (3). In Egypt, the estimated national prevalence of HCV is 14.7%, which is the highest in the world (4), and HCCs attributable to HCV account for around 50% of cases (5). Studies of the HCV genome confirmed that over 90% of Egyptian HCV isolates belong to genotype 4 (6).
The present study was conducted to characterize the molecular alterations in the insulin-like growth factor 1 receptor (IGF1R) pathway in a unified cohort of Egyptian HCCs complicating chronic HCV genotype 4. The IGF1R pathway was chosen as its activation plays an important role in almost every aspect of cancer development, including cell proliferation, neoplastic transformation, cancer cell survival, epithelial to mesenchymal transition (EMT), and metastasis (7) and that IGF1R overexpression correlated with aggressive phenotype in cancer, poor clinical outcome, and therapy resistance (8–10).
Aim
The present work aimed to identify the gene expression of the different subcellular components of the IGF1R pathway in Egyptian HCCs complicating chronic HCV genotype 4, followed by identification of the most frequently coregulated genes, correlation of gene expression with the different clinical and pathological patient and tumor characteristics, and determination of the activity status of involved pathways.
Materials
Two groups were included in the present study; the study group comprised 32 formalin-fixed, paraffin-embedded (FFPE) blocks from 32 Egyptian HCCs complicating chronic HCV genotype 4 etiology (shown as positive by HCV DNA PCR using Inno-LiPA I and II). Twenty-four out of 32 were segmentectomy specimens, while 8 out of 32 were liver explants, and the control group comprised six core biopsies from normal liver donors.
Inclusion criteria were HCV genotype 4 etiology and good quantity and quality of extracted ribonucleic acid (RNA) as measured by NanoDrop spectrophotometer (A260:A280 ratio 1.9:2.0 indicated pure RNA). Exclusion criteria included positive serology for anti-hepatitis B virus surface antibody and any HCC patient who has received previous neoadjuvant chemotherapy (including transarterial chemoembolization).
To accurately define our study population, further identification of other known hepatocarcinogens in the Egyptian environment was performed, including:
1) Hepatitis B core antigen (HBcAg) positivity by immunohistochemistry (IHC) (denoting occult hepatitis B virus [HBV] infection) was found in 7/29 of our HCC cases as evidenced by cytoplasmic positivity to the antibody.
2) Aflatoxin B1-DNA (AFB1-DNA) adduct positivity by IHC was found to be present in 16/29 of our cases as evidenced by nuclear positivity to the antibody.
Methods
The protocol of this study was approved by the Ethics committee of the Faculty of Medicine, University of Alexandria. Paraffin blocks of liver biopsies were obtained from the archives of the Pathology Department at the Faculty of Medicine, Alexandria University.
Clinical data were collected from the patients’ files including gender, age, comorbidities (diabetes mellitus and obesity), alfa-fetoprotein (AFP) levels, liver function tests (alanine aminotransferase [ALT], aspartate transaminase [AST], bilirubin, and albumin), hemoglobin concentration, and complete blood picture.
Gross pathologic examination of HCC tumors specifically addressed tumor size, multifocality, gross capsulation, gross necrosis, satellites, and bile production.
Histopathologic study using 5-μm-thick sections assessed pathologic tumor grade (was performed according to the Edmondson and Steiner grading system for HCC), histologic pattern, cell type (including giant cells and clear cells), and vascular invasion. The presence of dysplastic nodules outside HCC and pathological tumor-node-metastasis (pTNM) staging (according to the pTNM staging system of the American Joint Committee on Cancer 8th edition [AJCC]) was also noted.
Real-time quantitative reverse transcription PCR
Preparation of the paraffin blocks and obtaining tissue sections
1) By trimming off the excess paraffin from the edges of the specimen to facilitate deparaffinization.
2) Then two sections, 5 μm thick each, were cut from each block and placed in a 2 mL nuclease-free microfuge tube.
Total RNA was extracted from FFPE tumor sections using the RNeasy® FFPE kit for purification of total RNA from FFPE tissue sections (QIAGEN, Hilden, Germany, cat. no.: 73504) according to the manufacturer’s instructions. Its main principle is to remove all the paraffin, reverse formalin-induced cross-linking in the nucleic acid, and prevent any further degradation in the RNA that is known to be induced by formalin and finally to eliminate all genomic DNA. The concentration and purity of extracted RNA were determined using a NanoDrop ND-1000 spectrophotometer. Samples were accepted when the A260/A280 ratio (nucleic acid/protein absorbance) was ≥1.9. Reverse transcription of extracted RNA (0.5 µg [500 ng] of RNA was used for all experimental runs) to complementary DNA (cDNA) was done using RT2 First Strand Kit (QIAGEN, Hilden, Germany, cat. no.: 330401) according to the manufacturer’s instructions. RT2 SYBR Green ROX qPCR Mastermix (QIAGEN, Hilden, Germany, cat. no.: 330523) was added to the cDNA. The mixture was aliquoted into the 96 wells of the commercially available RT² Profiler™ PCR Array Human Insulin Signaling Pathway (PAHS-030ZA) (QIAGEN, Hilden, Germany, cat. no.: 330231) containing primer assays for 84 genes concerning the human insulin signaling pathway, including genes of the phosphatidylinositol-3-kinase (PI3K) and the mitogen-activated protein kinase (MAPK) signaling pathways that are the two major downstream signaling pathways for IGF1R signaling (11) and also genes involved in carbohydrate, lipid, and protein metabolism; transcription factors and transcription regulators; genes involved in cell proliferation, growth and differentiation (12); and five housekeeping genes (HKGs). In addition, there were one genomic DNA control, three reverse transcription controls, and three positive PCR controls (the full list of genes can be found in the supplementary file). PCR is then performed using the Applied Biosystems® 7500 Real-Time PCR system (ThermoFisher Scientific, Foster City, CA, USA). Finally, relative expression is determined using the ΔΔCT method using the RT2 Profiler PCR Array Data Analysis Webportal (at www.SABiosciences.com/pcrarraydataanalysis.php). Two HKGs with the lowest standard deviations and most stable expression across replicates were selected for data normalization: Beta-2-microglobulin (B2M) (H2) and hypoxanthine phosphoribosyl transferase 1 (HPRT1) (H4).
Fold regulation values for each of the 84 genes in HCC cases was determined by the Qiagen online data analysis Webportal (at www.SABiosciences.com/pcrarraydataanalysis.php). Then for each gene the percentages of cases showing upregulation, normal expression, and downregulation were determined. Determination of the significantly up- and downregulated genes in HCC cases using SPSS statistics software version 20.0 was carried out using nonparametric Mann-Whitney test. The most frequently coregulated genes were identified. Identification of genes significantly correlated with the different patient/tumor characteristics carried out using the Mann-Whitney test. Finally, active and inactive pathways in HCC from the present dataset were identified; an Excel file containing the gene symbols, fold regulation values, and the p values for all 84 tested genes was uploaded to the Ingenuity Pathway Analysis (IPA) software.
Results
The demographic data of the cases included are presented in Table I.
Gross pathological examination of HCC tumors
Tumor size ranged from 1.75 to 16.0 cm, with a median of 4.8 cm and a mean of 5.38 ± 3.46 cm standard deviation (SD; Tab. II).
Microscopic examination of HCC tumors
The most frequently encountered histologic patterns are displayed in Table III.
Twenty-six out of 32 cases showed combined histological patterns in the following frequencies (Tab. IV).
Frequency of different histopathological features of HCC tumors including Edmonson and Steiner tumor grades is shown in Tables V and VI, respectively.
Frequency of pTNM tumor stages is shown in Table VII.
| Number | % | |
|---|---|---|
| Gender | ||
| Male | 24 | 75.0 |
| Female | 8 | 25.0 |
| Age (years) | ||
| ≤60 | 22 | 68.8 |
| >60 | 10 | 31.3 |
| Min.–Max. | 47.0–67.0 | |
| Mean ± Standard deviation | 56.06 ± 5.49 | |
| Median | 56.0 | |
| Comorbidities | 17/25 | 53.1 |
| Alfa-fetoprotein (≥200 ng/mL) | 8/25 | 25.0 |
| Gross findings | Number | % |
|---|---|---|
| Gross capsulation (n = 32) | ||
| Absent | 6 | 18.8 |
| Present | 26 | 81.3 |
| Gross necrosis (n = 32) | ||
| Absent | 21 | 65.6 |
| Present | 11 | 34.4 |
| Multifocality (n = 32) | ||
| Absent | 4 | 12.5 |
| Present | 28 | 87.5 |
| Satellites (n = 32) | ||
| Absent | 6 | 18.8 |
| Present | 26 | 81.3 |
| Bile production (n = 32) | ||
| Absent | 15 | 46.9 |
| Present | 17 | 53.1 |
| Histologic pattern of HCC (n = 32) | Number | % |
|---|---|---|
| Trabecular | ||
| Absent | 3 | 9.4 |
| Present | 29 | 90.6 |
| Pseudoglandular | ||
| Absent | 12 | 37.5 |
| Present | 20 | 62.5 |
| Compact | ||
| Absent | 20 | 62.5 |
| Present | 12 | 37.5 |
HCC = hepatocellular carcinoma.
| HCCs with combined histologic pattern | Number (%) |
|---|---|
| Trabecular and pseudoglandular | 15 |
| Trabecular and compact | 6 |
| Trabecular, pseudoglandular, and compact | 3 |
| Glandular and compact | 2 |
| Total | 26 |
HCC = hepatocellular carcinoma.
| Histologic feature | Number | % |
|---|---|---|
| Giant cells (n = 32) | ||
| Absent | 17 | 53.1 |
| Present | 15 | 46.9 |
| Clear cells (n = 32) | ||
| Absent | 17 | 53.1 |
| Present | 15 | 46.9 |
| Vascular invasion (n = 32) | ||
| Absent | 11 | 34.4 |
| Present | 21 | 65.6 |
| Dysplastic nodules outside HCC (n= 32) | ||
| Absent | 13 | 40.6 |
| Present | 19 | 59.4 |
HCC = hepatocellular carcinoma.
| Grade | Number | % |
|---|---|---|
| Low grade (1 + 2) | 14 | 43.8 |
| Grade I | 1 | 3.1 |
| Grade II | 13 | 40.6 |
| High grade (3 + 4) | 18 | 56.3 |
| Grade III | 3 | 9.4 |
| Grade IV | 15 | 46.9 |
Significantly up- and downregulated genes
A total of 51 deregulated genes were found in the present cohort of HCCs. Fourteen genes were upregulated while 37 genes were downregulated. Statistically significant upregulation was observed in HCC patients, in comparison to the control group, in six genes, namely AEBP1, AKT2, FOS, PIK3R1, PRKCI, SHC1, where SHC1 was the most significantly overexpressed gene (p value: 0.001) (Tab. VIII, Fig. 1). SHC1, AEBP1, AKT2, and PKCI were the most frequently coregulated genes among the significantly upregulated genes.
| Stage | Number | % |
|---|---|---|
| Early stage (1 and 2) | 12 | 37.5 |
| Stage I | 5 | 15.6 |
| Stage II | 7 | 21.9 |
| Advanced stage (3) | 20 | 62.5 |
| Stage III | 20 | 62.5 |
| Stage IV | 0 | 0.0 |
AJCC = American Joint Committee on Cancer; TNM = tumor-node-metastasis.
| Position | Gene symbol | Fold regulation | p | Number | % |
|---|---|---|---|---|---|
| A04 | AEBP1 | 6.694 | 0.045* | 29 | 90.6 |
| A06 | AKT2 | 6.0287 | 0.013* | 26 | 81.3 |
| C01 | FOS | 3.9608 | 0.045* | 25 | 78.1 |
| E11 | PIK3R1 | 4.1363 | 0.020* | 25 | 78.1 |
| F05 | PRKCI | 7.7467 | 0.006* | 28 | 87.5 |
| G04 | SHC1 | 11.8326 | 0.001* | 30 | 93.8 |
p = p values for Mann-Whitney test for comparing between the two groups.
*Statistically significant at p ≤ 0.05.
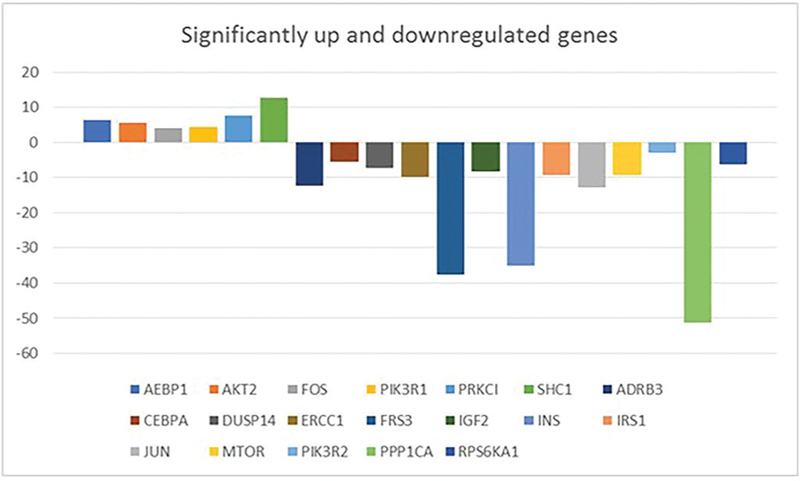
On the other hand, the frequency and magnitude of the downregulation events were greater than that of upregulation events (Tabs. VIII and IX). Thirteen genes, namely ADRB3, CEBPA, DUSP14, ERCC1, FRS3, IGF2, INS, IRS1, JUN, MTOR, PIK3R2, PPP1CA, and RPS6KA1, were significantly underexpressed in HCC patients relative to the control group, where PPP1CA and INS were the two most significantly underexpressed genes (p value: 0.002) (Tab. IX, Fig. 1). INS and PPP1CA were the most frequently coregulated genes among the significantly downregulated genes.
| Position | Gene symbol | Fold regulation | p | Number | % |
|---|---|---|---|---|---|
| A03 | ADRB3 | −11.3685 | 0.010* | 23 | 71.9 |
| B02 | CEBPA | −5.3981 | 0.041* | 22 | 68.8 |
| B07 | DUSP14 | −6.5717 | 0.016* | 23 | 71.9 |
| B10 | ERCC1 | −8.7853 | 0.037* | 22 | 68.8 |
| C03 | FRS3 | −35.5177 | 0.006* | 27 | 84.4 |
| D02 | IGF2 | −8.6788 | 0.013* | 25 | 78.1 |
| D04 | INS | −31.7678 | 0.002* | 30 | 93.8 |
| D07 | IRS1 | −8.0248 | 0.037* | 22 | 68.8 |
| D10 | JUN | −13.0027 | 0.020* | 23 | 71.9 |
| E04 | MTOR | −9.0478 | 0.045* | 26 | 81.3 |
| E12 | PIK3R2 | −2.8856 | 0.037* | 23 | 71.9 |
| F03 | PPP1CA | −46.4755 | 0.002* | 29 | 90.6 |
| F12 | RPS6KA1 | −5.653 | 0.025* | 24 | 75.0 |
p = p values for Mann-Whitney test for comparing between the two groups.
*Statistically significant at p ≤ 0.05.
VEGFA was downregulated (fold regulation of −3.2345) in 71.9% (23/32) of cases in the present study, although this did not reach statistical significance.
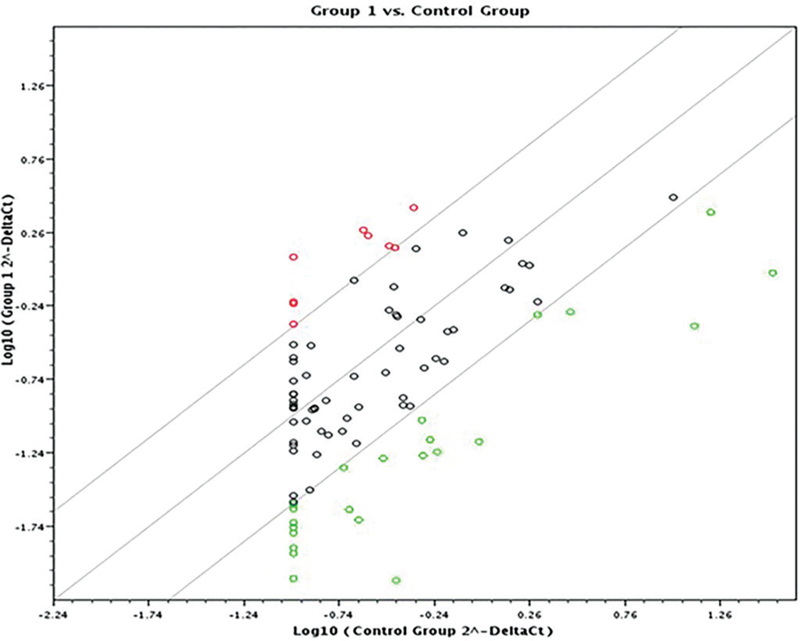
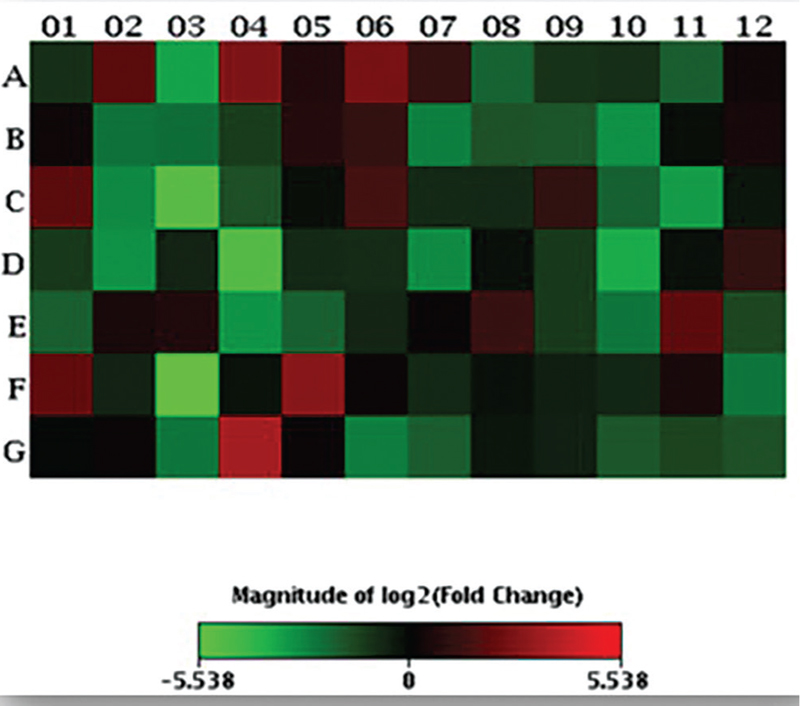
A scatter plot and a heat map were generated on the online data analysis web portal representing the distribution of average gene expression levels (Figs. 1 and 2 respectively).
Nonsupervised hierarchical clustering of all cases and controls according to their gene expression profile by the data analysis web portal
The included cases were a homogeneous population with no outstanding clustering as evidenced by the dendrograms. This further supports the study design as it included a specific cohort of HCCs: Egyptian ethnicity and HCV genotype 4 etiology.
Correlations between the gene expression levels and demographic data in HCC patients
Age: There were significant differences in three gene expression levels as regards age; AEBP1, AKT1, and FBP1 genes were significantly overexpressed in HCC patients ≤60 years of age compared to HCC patients >60 years of age (Tab. X).
Gender: DOK1 was significantly overexpressed in female patients (p = 0.037).
Stage: There were no statistically significant differences detected in the gene expression levels between early pTNM stage (pT1,2) and late stage (pT3,4) HCC tumors.
Grade: GSK3A was significantly overexpressed in low-grade tumors as compared to high-grade ones where it was normally expressed (p = 0.002)
| Gene symbol | Group 1
Age ≤60 years |
Group 1
Age >60 years |
p |
|---|---|---|---|
| Fold regulation | Fold regulation | ||
| AEBP1 | 9.6655 | 2.9833 | 0.040* |
| AKT1 | 2.3142 | −1.394 | 0.024* |
| FBP1 | 2.1679 | −2.1895 | 0.007* |
HCC = hepatocellular carcinoma.
p = p values for Mann-Whitney test for comparing between the two categories.
*Statistically significant at p ≤ 0.05.
Tumor size: Both CEBPB (p = 0.027) and GPD1 (p = 0.002) were significantly downregulated in tumors >5 cm.
Dysplastic nodules: No statistically significant difference was detected in the gene expression levels of any genes between cases who had dysplastic nodules outside tumor mass and those who did not have any (none were <0.05).
Satellite nodules: AEBP1 and FOS genes were upregulated in the absence of satellite nodules more than double their expression level in the cases which showed satellite nodules. MAPK1 gene was overexpressed in the presence of satellite nodules and normally expressed in their absence. PRKCZ and UCP1 genes were downregulated in the absence of satellite nodules and normally expressed in their presence (Tab. XI).
Multifocality: AEBP1 and FOS genes were upregulated in the absence of multifocal masses more than double their expression level in the cases which showed multifocal masses.
LDLR gene was overexpressed in the absence of multifocal masses and normally expressed in their presence (Tab. XII).
Gross capsulation: Both INSL3 (p = 0.043) and IRS1 (p = 0.030) genes were downregulated in the presence of gross capsulation.
Gross tumor necrosis: CAP1 (p = 0.041), GRB2 (p = 0.047), PRL (p = 0.024), and TG (p = 0.025) were downregulated in the presence of gross necrosis. PIK3R1 (p = 0.016) was upregulated in the absence of gross tumor necrosis.
| Satellite nodules | p | ||
|---|---|---|---|
| Absent (n = 6) | Present (n = 26) | ||
| AEBP1 | 17.44 | 5.37 | 0.004* |
| FOS | 13.21 | 3.00 | 0.007* |
| MAPK1 | −1.97 | 2.05 | 0.012* |
| PRKCZ | −2.63 | 1.42 | 0.026* |
| UCP1 | −9.60 | −1.98 | 0.014* |
HCC = hepatocellular carcinoma.
p = p values for Mann-Whitney test for comparing between the two categories.
*Statistically significant at p ≤ 0.05.
| Multifocality | p | ||
|---|---|---|---|
| Absent (n = 4) | Present (n = 28) | ||
| AEBP1 | 26.30 | 5.51 | 0.004* |
| FOS | 19.81 | 3.15 | 0.003* |
| LDLR | 7.12 | 1.88 | 0.035* |
HCC = hepatocellular carcinoma.
p = p values for Mann-Whitney test for comparing between the two categories.
*Statistically significant at p ≤ 0.05.
Giant cells: Both GSK3A (p = 0.015) and PCK2 (p = 0.030) were upregulated in the absence of giant cells. UCP1 (p = 0.033) was downregulated in tumors that showed giant cells.
Clear cells: CEBPB, HK2, GPD1, IGFBP1 and PIK3R2 were downregulated in all the studied cases, more so in tumors that showed clear cells than in tumors which had no clear cells (Tab. XIII).
Vascular invasion: NOS2 showed more significant downregulation in the absence of vascular invasion (p = 0.035).
HBcAg: RPS6KA1 was more significantly underexpressed in the presence of HBcAg (p = 0.034).
AFB1-DNA adduct positivity: AKT1 and MAPK1 were more significantly upregulated in the presence of aflatoxin as compared to negative cases (p = 0.019, 0.017 respectively). On the other hand, RPS6KA1 was more significantly underexpressed in the presence of AFB1 (p = 0.046).
Correlations between the gene expression levels and the different clinical characteristics of the tumors in HCC patients
Thrombocytopenia: In patients with a low platelet count, AKT1 was significantly upregulated (p = 0.038) while BCL2L1 was significantly downregulated (p = 0.045). VEGFA was more significantly underexpressed in the presence of normal platelet numbers (p = 0.038).
| Clear cells | p | ||
|---|---|---|---|
| Absent (n = 17) | Present (n = 15) | ||
| CEBPB | −2.34 | −12.28 | 0.010* |
| GPD1 | −1.29 | −2.64 | 0.040* |
| HK2 | −6.14 | −18.17 | 0.050* |
| IGFBP1 | 1.22 | −3.46 | 0.041* |
| PIK3R2 | −1.41 | −6.52 | 0.021* |
HCC = hepatocellular carcinoma.
p = p values for Mann-Whitney test for comparing between the two categories.
*Statistically significant at p ≤ 0.05.
Comorbidities: There was no statistically significant difference in the gene expression levels between patients who have comorbidities and those who do not have any.
Canonical pathway analysis predicted by IPA database
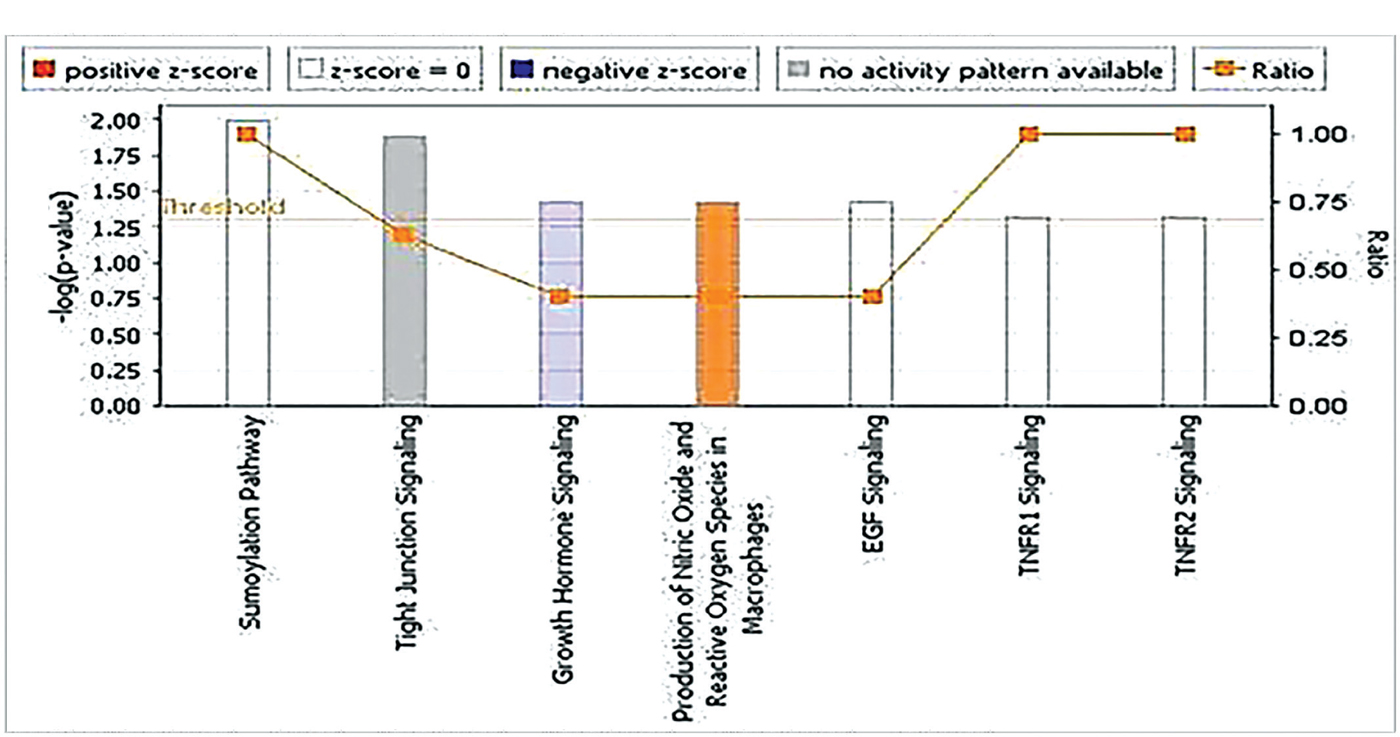
Nitric oxide and reactive oxygen species production pathway was significantly activated in the present cohort, while the growth hormone signaling pathway was significantly inactivated (Fig. 4).
Discussion
Different etiologic factors result in very diverse HCC molecular profiles between different regions of the world (13, 14). Understanding the heterogeneity of HCC is conducive to developing personalized therapy and identifying molecular biomarkers.
Most published work on molecular profiling of HCC was conducted on heterogeneous groups of tumors including different etiologies and ethnic backgrounds (15–19). Once HCC heterogeneity is accepted (20), studies have to be conducted on homogeneous samples to identify characteristic genetic alterations, if personalized management is the target.
HCC is known to be chemo- and radioresistant (21); therefore, identifying therapeutic vulnerabilities for molecular targeted drugs is in dire need. The present study measured levels of messenger RNA (mRNA) expression of 84 insulin signaling-related genes, including genes of the PI3K and the MAPK signaling pathways (11). Also included are genes involved in carbohydrate, lipid and protein metabolism, transcription factors and transcription regulators, genes involved in cell proliferation, growth, and differentiation (12).
Of the 84-cancer pathway-focused genes in this array, upregulation was observed in 14 genes, while 37 genes appeared to be downregulated in the tumor samples, for a total of 51 differentially regulated genes. These upregulated genes may be therapeutic targets for future trials, especially those for which molecularly targeted agents are available.
Among the significantly upregulated genes was c-FOS, a proto-oncogene which is a major nuclear target for signal transduction pathways, including MAPK pathway, and is involved in the regulation of cell growth, differentiation, and transformation (22,23). A novel selective C-FOS/AP-1 inhibitor T‐5224 was found to inhibit the invasion and migration of head and neck squamous cell carcinoma cells in vitro and prevented lymph node metastasis in head and neck cancer in an animal model (24).
Most of the significantly upregulated genes found in the present work were also found in common with prior HCC molecular studies; AKT2, C-FOS, PIK3R1, PRKCι, SHC1 were all found to be similarly significantly upregulated (25–30). AEBP1 is an exception; to the best of our knowledge, this is the first report to find this gene to be significantly and consistently (>90% of HCCs) upregulated in HCC. It is a transcriptional repressor that has been studied in great detail for its role in regulating adipogenesis (31). It has also been found to be a proinflammatory mediator (32). Finally, it has been shown that AEBP1 induces massive obesity in mice with targeted overexpression of AEBP1 in adipose tissue (33). Although its exact role in tumorigenesis is not clear (34), AEBP1 was found to be upregulated in many different cancers such as cervical cancer (35,36), primary glioblastoma multiforme (37), and human breast cancer cell lines (38). Ladha et al found that silencing AEBP1 expression and loss of its function lead to apoptosis in glioma cell lines, meaning that AEBP1 has an antiapoptotic function and is an important survival factor for tumor cells (34).
Two studies found that directly targeting AKT2 by miR-302b in human HCC cells suppresses cell proliferation, invasion, and metastasis in vitro and also inhibited HCC tumor growth in vivo (25,39). A recent preclinical in vitro and in vivo trial using a highly selective allosteric pan-AKT inhibitor in combination with sorafenib found that this combination yielded better outcomes and with minimal toxicity than sorafenib alone. The combination significantly reduced tumor growth, proliferation, and angiogenesis and increased apoptosis (40).
The PKCι blocking agent aurothiomalate (ATM) was found to negatively regulate EMT and invasion of HCC in immortalized murine hepatocytes and was found to inhibit proliferation and induce apoptosis in HepG2 cells (41). Also, a recent patent has been published on a novel drug targeting PKCι. It has been found to be effective at treating colon cancer cells having high level of expression of at least one aPKC (42). These can potentially be tested in preclinical trials as a method for treating HCCs overexpressing atypical PKCs.
Similarly, PKCι was upregulated in HCC in several studies involving different ethnic groups; two Chinese studies (Wang et al (43), Du et al (44)) and an American and Spanish study (Chiang et al (45)). Boyault et al in a French study attempted to classify HCC by unsupervised hierarchical clustering according to the tumors’ gene expression profiles by microarray; all three generated subgroups had upregulated PKCι (the HBV and non-HBV-related HCCs) (46).
In line with our findings, PIK3R1 was found to be upregulated in HCV-related HCC subgroups (45) and in HCC patients of Chinese ethnicity as well (47). In a recent study by He et al, PIK3R1 transfection by small interfering RNA significantly inhibited HCC cell migration and invasion (48). There are several PI3K inhibitors including both pan-PI3K and isoform-specific PI3K inhibitors that are still in early phase clinical trials (49).
The strongest associations between upregulated genes in the present study were between AEBP1, SHC1, AKT2, and PRKCi. Possibly, these associations can serve as a guide to molecularly targeted therapy combinations to cotarget the genes that were most frequently coregulated to maximize the benefits in HCC therapy. These potential therapeutic targets and combinations open several pipelines for future preclinical and clinical trials to be conducted for agents targeting those upregulated genes to test their efficacy and their potential use in HCV-related HCC treatment.
The 13 significantly downregulated genes included genes involved in carbohydrate, protein, and lipid metabolism, PI3K signaling target genes, MAPK signaling target genes, insulin receptor-associated proteins, genes involved in cell cycle, cell proliferation, growth factors and receptors, insulin gene and insulin receptor-associated proteins, primary insulin signaling target genes, transcription factors, and transcription regulators (12).
Two of those were tumor suppressor genes. CEBPA has been described as a tumor suppressor, leading to mitotic arrest through activation of p21 and repression of E2Fs and cyclin-dependent kinases (50). Its expression is downregulated in a number of cancers including lung (51), breast cancers (52), head and neck squamous cell carcinoma (53), and HCC. Its downregulation was associated with advanced HCC stages and poor survival (54,55).
Recently, a modified small activating RNA (saRNA) retained activation of CEBPA mRNA and downstream targets and inhibited growth of liver cancer cell lines in vitro (56). This novel drug has been encapsulated in a liposomal formulation for liver delivery in humans and is currently in a phase I clinical trial for patients with liver cancer and represents the first human study of a saRNA therapeutic (57).
FRS3 was significantly downregulated in our patient cohort. This is the first report of its significant downregulation in HCCs. Similar to our findings, FRS3 expression level has been found to be downregulated in brain and lung cancer cell lines. It constitutively binds to EGFR regardless of EGF stimulation and inhibits EGF-induced cell transformation and proliferation (58). It serves as a tumor suppressor in non-small cell lung cancer (NSCLC) (59).
In our work, excision repair cross-complementing rodent repair deficiency, complementation group 1 (ERCC1) was significantly downregulated. It was also found to be downregulated in Chinese (47) and Turkish (60) HCC cohorts as well. Loss of ERCC1 expression in hepatocytes may be one of the leading factors for genetic instability, and thus of tumorigenesis (61). Impaired nucleotide excision repair pathway, in which ERCC1 plays a major role, leads to reduced DNA repair capability and increases DNA adduct levels and genomic instability that in turn could lead to a more malignant phenotype (62). This is not limited to HCCs, but also is a finding in other types of cancer including head and neck squamous cell carcinoma (63).
Gene expression patterns can be used to define the suitable chemotherapeutic agent employed in any given case. ERCC1 overexpression could be associated with resistance to cisplatin-based therapy in HCC, similar to what is observed in NSCLC (64,65).
VEGFA was found to be downregulated in the present study in 71.9% of cases. Several retrospective studies suggested that tumors with a VEGFA amplification respond better to sorafenib, the only approved first-line treatment of advanced HCC (66), suggesting that further studies of currently used molecularly targeted therapies in Egyptian HCV-related HCC patients are needed, and search for molecular biomarkers of response to therapy with prospective validation of the role of VEGFA expression level to predict response to therapy.
One of the points of strength in our study is that we included HCC samples resulting from a single etiologic agent only (HCV genotype 4-related HCC). Also, the present study unified the controls used in the analysis as we used healthy liver donors as the control group. On the other hand, in the vast majority of studies paired adjacent cirrhotic liver tissues to the HCC tumor masses were used as controls (30,45,67–69). This complicates the interpretation of results since the gene expression patterns in the nontumor cirrhotic liver samples from different patients can vary significantly, affected by the viral infection and degree of fibrosis. Thus, the tumor-specific variation in the expression patterns could not be distinguished from variation due to differences in the corresponding nontumor liver samples (70).
In our cohort, two HKGs – B2M and HPRT1 – were used for data normalization. In a study performed to search for appropriate HKGs to be used for data normalization in HCV-induced HCC specifically. Two of the most commonly used HKGs, which were also included in the standard catalogued 5 HKGs set in the present study’s array, namely glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and beta-actin (ACTB), were found to be deregulated in HCV-induced HCC (71). Thus, using them for normalization would have strong effects on the extent of differential expression of genes, leading to misinterpretation of results. This was the exact same case in our study for ACTB and GAPDH, which were found to be unstable and had a wide SD across samples.
A few weaknesses of this study that we recognize are, firstly, the fact that we were studying patients who already had developed disease at a single point in time. Also, as in most studies on HCC, tumor samples have been collected during resection. It could be expected that in a more clinically advanced context with tumor progression, increased crosstalk among pathways and chromosomal instability would contribute to additional signaling deregulation. These samples do not represent the tumors of patients with advanced stage disease, who do not undergo surgery. However, as HCC diagnosis at present is not tissue based, obtaining tissue samples from late cases is not feasible at present.
Conclusions
The significantly up- and downregulated genes identified in the present study can serve as potential therapeutic targets for HCV-related HCCs.
In the present study, the insulin pathway genes including IGF1R, IGF2, INS, INSR, and IRS1 were found to be downregulated and the IGF pathway was found to be significantly inactive, in the present work, which is in concordance with the findings of Zucman-Rossi et al (72). Therefore, drugs targeting the IGF1R pathway may not be suitable for this specific subset of HCV-related HCCs.
Although NOS2 gene expression was unchanged in our cohort, IPA revealed that nitric oxide pathway and reactive oxygen species production in HCV-related HCCs were significantly activated.
The gene expression pattern of each HCC tumor seems to provide a distinctive molecular portrait of that tumor, as no gene was 100% up- or downregulated in the present cohort. Personalized medicine mandates the knowledge of the specific genetic profile of each HCC tumor, which is currently not feasible due to costs associated especially in developing countries where most of the HCC burden lies. Therefore, the present study fills a void in this respect – identifying specific morphologic and clinical patient/tumor characteristics, which significantly correlate with underlying molecular alterations. This may be used as a guide for tailoring molecularly targeted therapies.
We confirm the findings of a previous study that was performed to search for appropriate HKGs to be used for data normalization in HCV-induced HCC specifically. Two of the most commonly used HKGs – ACTB and GAPDH – were found to be unstable and had a wide SD in our study and theirs; therefore these shouldn’t be used for normalization. We suggest B2M and HPRT1 to be used instead, as they had more stable levels of expression.
Disclosures
Financial support: The present study was fully funded by a grant from the “Alexandria Regional Center for Women’s Health and Development,” Alexandria, Egypt (this grant covered all expenses needed for provision of all experimental kits).
Conflict of interest: All the authors declare no conflict of interest.
References
- 1. International Agency for Research on Cancer (IARC). Globocan 2012: Estimated cancer incidence, mortality and prevalence worldwide in 2012. France: IARC; 2012.
- 2. Shi J, Zhu L, Liu S, Xie WF. A meta-analysis of case-control studies on the combined effect of hepatitis B and C virus infections in causing hepatocellular carcinoma in China. Br J Cancer. 2005;92(3):607–612.
- 3. Llovet JM, Villanueva A, Lachenmayer A, Finn RS. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat Rev Clin Oncol. 2015;12(7):408–424.
- 4. El-zanaty F, Way A. Egypt Demographic and Health Survey 2008. Cairo, Egypt: Ministry of Health, El-Zanaty and Associates, and Macro International; 2009.
- 5. Maucort-Boulch D, de Martel C, Franceschi S, Plummer M. Fraction and incidence of liver cancer attributable to hepatitis B and C viruses worldwide. Int J Cancer. 2018;142(12):2471-7.
- 6. Ray SC, Arthur RR, Carella A, Bukh J, Thomas DL. Genetic epidemiology of hepatitis C virus throughout Egypt. J Infect Dis. 2000;182(3):698–707.
- 7. Wang Y, Sun Y. Insulin-like growth factor receptor-1 as an anti-cancer target: blocking transformation and inducing apoptosis. Curr Cancer Drug Targets. 2002;2(3):191–207.
- 8. Christopoulos PF, Msaouel P, Koutsilieris M. The role of the insulin-like growth factor-1 system in breast cancer. Mol Cancer. 2015;14(1):43.
- 9. Papa V, Gliozzo B, Clark GM, et al. Insulin-like growth factor-I receptors are overexpressed and predict a low risk in human breast cancer. Cancer Res. 1993;53(16):3736–3740.
- 10. Jones R, Campbell C, Gunther E, et al. Transgenic overexpression of IGF-IR disrupts mammary ductal morphogenesis and induces tumor formation. Oncogene. 2007;26(11):1636–1644.
- 11. Samani AA, Yakar S, LeRoith D, Brodt P. The role of the IGF system in cancer growth and metastasis: overview and recent insights. Endocr Rev. 2007;28(1):20–47.
- 12. Vescovo T, Refolo G, Vitagliano G, Fimia GM, Piacentini M. Molecular mechanisms of hepatitis c virus–induced hepatocellular carcinoma. Clin Microbiol Infect. 2016;22(10):853–861.
- 13. Patel A, Sun W. Molecular targeted therapy in hepatocellular carcinoma: from biology to clinical practice and future. Curr Treat Opt Oncol. 2014;15(3):380–394.
- 14. Lu L-C, Hsu C-H, Hsu C, Cheng A-L. Tumor heterogeneity in hepatocellular carcinoma: facing the challenges. Liver Cancer. 2016;5(2):128–138.
- 15. Luo JH, Ren B, Keryanov S, et al. Transcriptomic and genomic analysis of human hepatocellular carcinomas and hepatoblastomas. Hepatology. 2006;44(4):1012–1024.
- 16. Lee JS, Chu IS, Heo J, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40(3):667–676.
- 17. Shirota Y, Kaneko S, Honda M, Kawai HF, Kobayashi K. Identification of differentially expressed genes in hepatocellular carcinoma with cDNA microarrays. Hepatology. 2001;33(4):832–840.
- 18. Tackels-Horne D, Goodman MD, Williams AJ, et al. Identification of differentially expressed genes in hepatocellular carcinoma and metastatic liver tumors by oligonucleotide expression profiling. Cancer. 2001;92(2):395–405.
- 19. Patil MA, Chua M-S, Pan K-H, et al. An integrated data analysis approach to characterize genes highly expressed in hepatocellular carcinoma. Oncogene. 2005;24(23):3737–3747.
- 20. Guimei M, Baddour N, ElKaffash D, Abdou L, Taher Y. Gremlin in the pathogenesis of hepatocellular carcinoma complicating chronic hepatitis C: an immunohistochemical and PCR study of human liver biopsies. BMC Res Notes. 2012;5(1):390.
- 21. Huynh H. Molecularly targeted therapy in hepatocellular carcinoma. Biochem Pharmacol. 2010;80(5):550–560.
- 22. Saez E, Rutberg SE, Mueller E, et al. C-fos is required for malignant progression of skin tumors. Cell. 1995;82(5):721–732.
- 23. Yuen MF, Wu PC, Lai VCH, Lau JYN, Lai CL. Expression of c-Myc, c-Fos, and c-Jun in hepatocellular carcinoma. Cancer. 2001; 91(1):106–112.
- 24. Kamide D, Yamashita T, Araki K, et al. Selective activator protein-1 inhibitor t-5224 prevents lymph node metastasis in an oral cancer model. Cancer Sci. 2016;107(5):666–673.
- 25. Wang L, Yao J, Zhang X, et al. Mirna-302b suppresses human hepatocellular carcinoma by targeting Akt2. Mol Cancer Res. 2014;12(2):190–202.
- 26. Zhang Y, Guo X, Yang M, Yu L, Li Z, Lin N. Identification of AKT kinases as unfavorable prognostic factors for hepatocellular carcinoma by a combination of expression profile, interaction network analysis and clinical validation. Mol BioSyst. 2014;10(2):215–222.
- 27. Kurokawa Y, Matoba R, Takemasa I, et al. Molecular features of non-B, non-C hepatocellular carcinoma: a PCR-array gene expression profiling study. J Hepatol. 2003;39(6):1004–1012.
- 28. Huang C-Y, Huang X-P, Zhu J-Y, et al. miR-128-3p suppresses hepatocellular carcinoma proliferation by regulating PIK3R1 and is correlated with the prognosis of HCC patients. Oncol Rep. 2015;33(6):2889–2898.
- 29. Wang JM, Li Q, Du GS, Lu JX, Zou SQ. Significance and expression of atypical protein kinase C-iota in human hepatocellular carcinoma. J Surg Res. 2009;154(1):143–149.
- 30. Chen X, Cheung ST, So S, et al. Gene expression patterns in human liver cancers. Mol Biol Cell. 2002;13(6):1929–1939.
- 31. Ro H-S, Roncari D. The C/EBP-binding region and adjacent sites regulate expression of the adipose p2 gene in human preadipocytes. Mol Cell Biol. 1991;11(4):2303–2306.
- 32. Majdalawieh A, Ro HS. Regulation of IkappaBalpha function and NF-kappaB signaling: AEBP1 is a novel proinflammatory mediator in macrophages. Mediators Inflamm. 2010;2010:823–821.
- 33. Zhang L, Reidy SP, Nicholson TE, et al. The role of AEBP1 in sex-specific diet-induced obesity. Mol Med. 2005;11(1–12):39.
- 34. Ladha J, Sinha S, Bhat V, Donakonda S, Rao SM. Identification of genomic targets of transcription factor AEBP1 and its role in survival of glioma cells. Mol Cancer Res. 2012;10(8):1039–1051.
- 35. Han S-J, Cho YL, Nam GH, Kim CK, Seo J-S, Ahn WS. CDNA microarray analysis of gene expression profiles associated with cervical cancer. Cancer Res Treat. 2003;35(5):451–459.
- 36. Ahn WS, Bae SM, Lee JM, et al. Searching for pathogenic gene functions to cervical cancer. Gynecol Oncol. 2004;93(1):41–48.
- 37. Reddy SP, Britto R, Vinnakota K, et al. Novel glioblastoma markers with diagnostic and prognostic value identified through transcriptome analysis. Clin Cancer Res. 2008;14(10):2978–2987.
- 38. Grigoriadis A, Mackay A, Reis-Filho JS, et al. Establishment of the epithelial-specific transcriptome of normal and malignant human breast cells based on MPSS and array expression data. Breast Cancer Res. 2006;8(5):R56.
- 39. Wang L, Yao J, Sun H, et al. miR-302b suppresses cell invasion and metastasis by directly targeting AKT2 in human hepatocellular carcinoma cells. Tumor Biol. 2016;37(1):847–855.
- 40. Jilkova ZM, Kuyucu AZ, Kurma K, et al. Combination of AKT inhibitor ARQ 092 and sorafenib potentiates inhibition of tumor progression in cirrhotic rat model of hepatocellular carcinoma. Oncotarget. 2018;9(13):11145–11158.
- 41. Ma C, Yang Y, Wang J, et al. The aPKCι blocking agent atm negatively regulates EMT and invasion of hepatocellular carcinoma. Cell Death Dis. 2014;5(3):e1129.
- 42. Acevedo-Duncan M. Method of treating colorectal cancers using a PKC inhibitor. In: Google Patents; 2018.
- 43. Wang J-M, Li Q, Du G-S, Lu J-X, Zou S-Q. Significance and expression of atypical protein kinase c-ι in human hepatocellular carcinoma. J Surg Res. 2009;154(1):143–149.
- 44. Du G-S, Wang J-M, Lu J-X, et al. Expression of P-aPKC-ι, E-cadherin, and β-catenin related to invasion and metastasis in hepatocellular carcinoma. Ann Surg Oncol. 2009;16(6):1578–1586.
- 45. Chiang DY, Villanueva A, Hoshida Y, et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68(16):6779–6788.
- 46. Boyault S, Rickman DS, De Reyniès A, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45(1):42–52.
- 47. De Yun F, Hui Z, Tan Y, Cheng RX. Effect of phosphorylation of MAPK and Stat3 and expression of c-fos and c-jun proteins on hepatocarcinogenesis and their clinical significance. World J Gastroenterol. 2001;7(1):33–36.
- 48. He S, Zhang J, Zhang W, Chen F, Luo R. Foxa1 inhibits hepatocellular carcinoma progression by suppressing PIK3R1 expression in male patients. J Exp Clin Cancer Res. 2017;36(1):175.
- 49. Rodon J, Tabernero J. Improving the armamentarium of PI3K inhibitors with isoform-selective agents: a new light in the darkness. Cancer Discov. 2017;7(7):666–669.
- 50. Schuster MB, Porse BT. C/EBPalpha: a tumour suppressor in multiple tissues? Biochim Biophys Acta. 2006;1766(1):88–103.
- 51. Costa DB, Li S, Kocher O, et al. Immunohistochemical analysis of C/EBPalpha in non-small cell lung cancer reveals frequent down-regulation in stage II and IIIA tumors: a correlative study of e3590. Lung Cancer (Amsterdam, Netherlands). 2007; 56(1):97–103.
- 52. Gery S, Tanosaki S, Bose S, Bose N, Vadgama J, Koeffler HP. Down-regulation and growth inhibitory role of C/EBPalpha in breast cancer. Clin Cancer Res. 2005;11(9):3184–3190.
- 53. Bennett KL, Hackanson B, Smith LT, et al. Tumor suppressor activity of CCAAT/enhancer binding protein alpha is epigenetically down-regulated in head and neck squamous cell carcinoma. Cancer Res. 2007;67(10):4657–4664.
- 54. Tseng HH, Hwang YH, Yeh KT, Chang JG, Chen YL, Yu HS. Reduced expression of C/EBPalpha protein in hepatocellular carcinoma is associated with advanced tumor stage and shortened patient survival. J Cancer Res Clin Oncol. 2009;135(2): 241–247.
- 55. Tomizawa M, Watanabe K, Saisho H, Nakagawara A, Tagawa M. Down-regulated expression of the CCAAT/enhancer binding protein alpha and beta genes in human hepatocellular carcinoma: a possible prognostic marker. Anticancer Res. 2003;23(1a):351–354.
- 56. Voutila J, Reebye V, Roberts TC, et al. Development and mechanism of small activating RNA targeting CEBPA, a novel therapeutic in clinical trials for liver cancer. Mol Ther. 2017;25(12): 2705–2714.
- 57. Reebye V, Huang KW. Gene activation of CEBPA using saRNA: preclinical studies of the first in human saRNA drug candidate for liver cancer. 2018;37(24):3216-28.
- 58. Huang L, Watanabe M, Chikamori M, et al. Unique role of SNT-2/FRS2β/FRS3 docking/adaptor protein for negative regulation in EGF receptor tyrosine kinase signaling pathways. Oncogene. 2006;25(49):6457.
- 59. Iejima D, Minegishi Y, Takenaka K, et al. FRS2β, a potential prognostic gene for non-small cell lung cancer, encodes a feedback inhibitor of EGF receptor family members by ERK binding. Oncogene. 2010;29(21):3087.
- 60. Turhal N, Bas E, Er O, et al. ERCC1 is not expressed in hepatocellular cancer: a Turkish oncology group, gastrointestinal oncology subgroup study. J Buon. 2010;15(4):794–796.
- 61. Cheng L, Spitz MR, Hong WK, Wei Q. Reduced expression levels of nucleotide excision repair genes in lung cancer: a case-control analysis. Carcinogenesis. 2000;21(8):1527–1530.
- 62. Simon GR, Sharma S, Cantor A, Smith P, Bepler G. ERCC1 expression is a predictor of survival in resected patients with non-small cell lung cancer. Chest. 2005;127(3):978–983.
- 63. Cheng L, Sturgis EM, Eicher SA, Spitz MR, Wei Q. Expression of nucleotide excision repair genes and the risk for squamous cell carcinoma of the head and neck. Cancer. 2002;94(2):393–397.
- 64. Ueda S, Shirabe K, Morita K, et al. Evaluation of ERCC1 expression for cisplatin sensitivity in human hepatocellular carcinoma. Ann Surg Oncol. 2011;18(4):1204–1211.
- 65. Fautrel A, Andrieux L, Musso O, Boudjema K, Guillouzo A, Langouët S: Overexpression of the two nucleotide excision repair genes ERCC1 and XPC in human hepatocellular carcinoma. J Hepatol. 2005;43(2):288–293.
- 66. Lopez PM, Patel P, Uva P, Villanueva A, Llovet JM. Current management of liver cancer. Eur J Cancer Suppl. 2007;5(5):444–446.
- 67. Xu XR, Huang J, Xu ZG, Qian BZ, et al. Insight into hepatocellular carcinogenesis at transcriptome level by comparing gene expression profiles of hepatocellular carcinoma with those of corresponding noncancerous liver. Proc Natl Acad Sci USA. 2001;98(26):15089–15094.
- 68. Boyault S, Rickman DS, de Reynies A, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45(1):42–52.
- 69. Chuma M, Sakamoto M, Yamazaki K, et al. Expression profiling in multistage hepatocarcinogenesis: identification of HSP70 as a molecular marker of early hepatocellular carcinoma. Hepatology. 2003;37(1):198–207.
- 70. Makowska Z, Boldanova T, Adametz D, et al. Gene expression analysis of biopsy samples reveals critical limitations of transcriptome-based molecular classifications of hepatocellular carcinoma. J Pathol Clin Res. 2016;2(2):80–92.
- 71. Waxman S, Wurmbach E. De-regulation of common housekeeping genes in hepatocellular carcinoma. BMC Genomics. 2007;8:243.
- 72. Zucman-Rossi J, Villanueva A, Nault J-C, Llovet JM. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology. 2015;149(5):1226–1239. e1224.